Genome-Wide Identification and Characterization of CDPK Family Reveal Their Involvements in Growth and Development and Abiotic Stress in Sweet Potato and Its Two Diploid Relatives
- PMID: 35328509
- PMCID: PMC8952862
- DOI: 10.3390/ijms23063088
Genome-Wide Identification and Characterization of CDPK Family Reveal Their Involvements in Growth and Development and Abiotic Stress in Sweet Potato and Its Two Diploid Relatives
Abstract
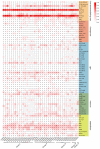
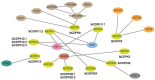
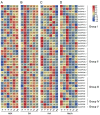
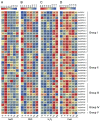
Calcium-dependent protein kinase (CDPKs) is one of the calcium-sensing proteins in plants. They are likely to play important roles in growth and development and abiotic stress responses. However, these functions have not been explored in sweet potato. In this study, we identified 39 CDPKs in cultivated hexaploid sweet potato (Ipomoea batatas, 2n = 6x = 90), 35 CDPKs in diploid relative Ipomoea trifida (2n = 2x = 30), and 35 CDPKs in Ipomoea triloba (2n = 2x = 30) via genome structure analysis and phylogenetic characterization, respectively. The protein physiological property, chromosome localization, phylogenetic relationship, gene structure, promoter cis-acting regulatory elements, and protein interaction network were systematically investigated to explore the possible roles of homologous CDPKs in the growth and development and abiotic stress responses of sweet potato. The expression profiles of the identified CDPKs in different tissues and treatments revealed tissue specificity and various expression patterns in sweet potato and its two diploid relatives, supporting the difference in the evolutionary trajectories of hexaploid sweet potato. These results are a critical first step in understanding the functions of sweet potato CDPK genes and provide more candidate genes for improving yield and abiotic stress tolerance in cultivated sweet potato.
Keywords: CDPK; Ipomoea batatas; Ipomoea trifida; Ipomoea triloba; abiotic stress; hormone treatment; tissue specificity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Genome-Wide Identification and Expression Analysis of ACTIN Family Genes in the Sweet Potato and Its Two Diploid Relatives.Int J Mol Sci. 2023 Jun 30;24(13):10930. doi: 10.3390/ijms241310930. Int J Mol Sci. 2023. PMID: 37446107 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of JAZ Family Involved in Hormone and Abiotic Stress in Sweet Potato and Its Two Diploid Relatives.Int J Mol Sci. 2021 Sep 10;22(18):9786. doi: 10.3390/ijms22189786. Int J Mol Sci. 2021. PMID: 34575953 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of SWEET Family Genes in Sweet Potato and Its Two Diploid Relatives.Int J Mol Sci. 2022 Dec 13;23(24):15848. doi: 10.3390/ijms232415848. Int J Mol Sci. 2022. PMID: 36555491 Free PMC article.
-
Current status in whole genome sequencing and analysis of Ipomoea spp.Plant Cell Rep. 2019 Nov;38(11):1365-1371. doi: 10.1007/s00299-019-02464-4. Epub 2019 Aug 29. Plant Cell Rep. 2019. PMID: 31468128 Free PMC article. Review.
-
Multiple biological functions of sporamin related to stress tolerance in sweet potato (Ipomoea batatas Lam).Biotechnol Adv. 2012 Nov-Dec;30(6):1309-17. doi: 10.1016/j.biotechadv.2012.01.022. Epub 2012 Jan 28. Biotechnol Adv. 2012. PMID: 22306516 Review.
Cited by
-
Identification of SikCDPK family genes to low-temperature by RNA-seq approaches and functional analysis of SikCDPK1 in Saussurea involucrata (Kar. & Kir.).Front Plant Sci. 2024 Sep 30;15:1436651. doi: 10.3389/fpls.2024.1436651. eCollection 2024. Front Plant Sci. 2024. PMID: 39403619 Free PMC article.
-
Genome-Wide Identification and Characterization of CDPK Gene Family in Cultivated Peanut (Arachis hypogaea L.) Reveal Their Potential Roles in Response to Ca Deficiency.Cells. 2023 Nov 21;12(23):2676. doi: 10.3390/cells12232676. Cells. 2023. PMID: 38067104 Free PMC article.
-
Genome-wide characterization and analysis of Golden 2-Like transcription factors related to leaf chlorophyll synthesis in diploid and triploid Eucalyptus urophylla.Front Plant Sci. 2022 Jul 28;13:952877. doi: 10.3389/fpls.2022.952877. eCollection 2022. Front Plant Sci. 2022. PMID: 35968152 Free PMC article.
-
Identification of the CDPK gene family in patchouli and functional analysis in response to continuous cropping stress.Front Plant Sci. 2023 Nov 22;14:1300073. doi: 10.3389/fpls.2023.1300073. eCollection 2023. Front Plant Sci. 2023. PMID: 38078089 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of ACTIN Family Genes in the Sweet Potato and Its Two Diploid Relatives.Int J Mol Sci. 2023 Jun 30;24(13):10930. doi: 10.3390/ijms241310930. Int J Mol Sci. 2023. PMID: 37446107 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

