A Homozygous Deep Intronic Variant Causes Von Willebrand Factor Deficiency and Lack of Endothelial-Specific Secretory Organelles, Weibel-Palade Bodies
- PMID: 35328514
- PMCID: PMC8950443
- DOI: 10.3390/ijms23063095
A Homozygous Deep Intronic Variant Causes Von Willebrand Factor Deficiency and Lack of Endothelial-Specific Secretory Organelles, Weibel-Palade Bodies
Abstract
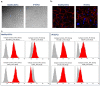
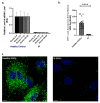
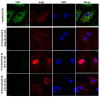
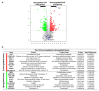
A type 3 von Willebrand disease (VWD) index patient (IP) remains mutation-negative after completion of the conventional diagnostic analysis, including multiplex ligation-dependent probe amplification and sequencing of the promoter, exons, and flanking intronic regions of the VWF gene (VWF). In this study, we intended to elucidate causative mutation through next-generation sequencing (NGS) of the whole VWF (including complete intronic region), mRNA analysis, and study of the patient-derived endothelial colony-forming cells (ECFCs). The NGS revealed a variant in the intronic region of VWF (997 + 118 T > G in intron 8), for the first time. The bioinformatics assessments (e.g., SpliceAl) predicted this variant creates a new donor splice site (ss), which could outcompete the consensus 5′ donor ss at exon/intron 8. This would lead to an aberrant mRNA that contains a premature stop codon, targeting it to nonsense-mediated mRNA decay. The subsequent quantitative real-time PCR confirmed the virtual absence of VWF mRNA in IP ECFCs. Additionally, the IP ECFCs demonstrated a considerable reduction in VWF secretion (~6% of healthy donors), and they were devoid of endothelial-specific secretory organelles, Weibel−Palade bodies. Our findings underline the potential of NGS in conjunction with RNA analysis and patient-derived cell studies for genetic diagnosis of mutation-negative type 3 VWD patients.
Keywords: ECFCs; Weibel–Palade bodies; angiopoietin-2; deep intronic mutation; next-generation sequencing; von Willebrand disease; von Willebrand factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Re-establishment of VWF-dependent Weibel-Palade bodies in VWD endothelial cells.Blood. 2005 Jan 1;105(1):145-52. doi: 10.1182/blood-2004-02-0464. Epub 2004 Aug 26. Blood. 2005. PMID: 15331450 Free PMC article.
-
The severe von Willebrand disease variant p.M771V leads to impaired anterograde trafficking of von Willebrand factor in patient-derived and base-edited endothelial colony-forming cells.J Thromb Haemost. 2025 Feb;23(2):466-479. doi: 10.1016/j.jtha.2024.10.023. Epub 2024 Nov 6. J Thromb Haemost. 2025. PMID: 39510415
-
von Willebrand disease-specific defects and proteomic signatures in endothelial colony-forming cells.J Thromb Haemost. 2025 Aug;23(8):2634-2650. doi: 10.1016/j.jtha.2025.04.024. Epub 2025 May 6. J Thromb Haemost. 2025. PMID: 40339962
-
Von Willebrand disease and Weibel-Palade bodies.Hamostaseologie. 2010 Aug;30(3):150-5. Hamostaseologie. 2010. PMID: 20680229 Review.
-
Emerging mechanisms to modulate VWF release from endothelial cells.Int J Biochem Cell Biol. 2021 Feb;131:105900. doi: 10.1016/j.biocel.2020.105900. Epub 2020 Dec 7. Int J Biochem Cell Biol. 2021. PMID: 33301925 Review.
Cited by
-
Genetic analysis using long-read sequencing to overcome the difficulties in VWF gene.Res Pract Thromb Haemost. 2025 May 17;9(4):102888. doi: 10.1016/j.rpth.2025.102888. eCollection 2025 May. Res Pract Thromb Haemost. 2025. PMID: 40529342 Free PMC article.
-
Interplay between circulating von Willebrand factor and neutrophils: implications for inflammation, neutrophil function, and von Willebrand factor clearance.Haematologica. 2025 Aug 1;110(8):1786-1797. doi: 10.3324/haematol.2023.284919. Epub 2025 Jan 9. Haematologica. 2025. PMID: 39781642 Free PMC article.
References
-
- James P.D., Connell N.T., Ameer B., Di Paola J., Eikenboom J., Giraud N., Haberichter S., Jacobs-Pratt V., Konkle B., McLintock C., et al. ASH ISTH NHF WFH 2021 guidelines on the diagnosis of von Willebrand disease. Blood Adv. 2021;5:280–300. doi: 10.1182/bloodadvances.2020003265. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

