Structural Consequence of Non-Synonymous Single-Nucleotide Variants in the N-Terminal Domain of LIS1
- PMID: 35328531
- PMCID: PMC8955593
- DOI: 10.3390/ijms23063109
Structural Consequence of Non-Synonymous Single-Nucleotide Variants in the N-Terminal Domain of LIS1
Abstract
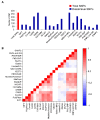
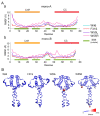
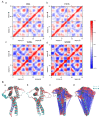
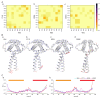
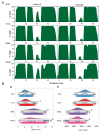
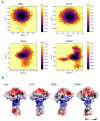
Disruptive neuronal migration during early brain development causes severe brain malformation. Characterized by mislocalization of cortical neurons, this condition is a result of the loss of function of migration regulating genes. One known neuronal migration disorder is lissencephaly (LIS), which is caused by deletions or mutations of the LIS1 (PAFAH1B1) gene that has been implicated in regulating the microtubule motor protein cytoplasmic dynein. Although this class of diseases has recently received considerable attention, the roles of non-synonymous polymorphisms (nsSNPs) in LIS1 on lissencephaly progression remain elusive. Therefore, the present study employed combined bioinformatics and molecular modeling approach to identify potential damaging nsSNPs in the LIS1 gene and provide atomic insight into their roles in LIS1 loss of function. Using this approach, we identified three high-risk nsSNPs, including rs121434486 (F31S), rs587784254 (W55R), and rs757993270 (W55L) in the LIS1 gene, which are located on the N-terminal domain of LIS1. Molecular dynamics simulation highlighted that all variants decreased helical conformation, increased the intermonomeric distance, and thus disrupted intermonomeric contacts in the LIS1 dimer. Furthermore, the presence of variants also caused a loss of positive electrostatic potential and reduced dimer binding potential. Since self-dimerization is an essential aspect of LIS1 to recruit interacting partners, thus these variants are associated with the loss of LIS1 functions. As a corollary, these findings may further provide critical insights on the roles of LIS1 variants in brain malformation.
Keywords: LIS1; lissencephaly; molecular dynamics simulation; single nucleotide polymorphisms; variant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
LIS1 functions in normal development and disease.Curr Opin Neurobiol. 2013 Dec;23(6):951-6. doi: 10.1016/j.conb.2013.08.001. Epub 2013 Aug 23. Curr Opin Neurobiol. 2013. PMID: 23973156 Review.
-
[Molecular mechanism of lissencephaly--how LIS1 and NDEL1 regulate cytoplasmic dynein?].Brain Nerve. 2008 Apr;60(4):375-81. Brain Nerve. 2008. PMID: 18421979 Review. Japanese.
-
Inhibition of calpain increases LIS1 expression and partially rescues in vivo phenotypes in a mouse model of lissencephaly.Nat Med. 2009 Oct;15(10):1202-7. doi: 10.1038/nm.2023. Epub 2009 Sep 6. Nat Med. 2009. PMID: 19734909 Free PMC article.
-
LIS1 controls mitosis and mitotic spindle organization via the LIS1-NDEL1-dynein complex.Hum Mol Genet. 2014 Jan 15;23(2):449-66. doi: 10.1093/hmg/ddt436. Epub 2013 Sep 12. Hum Mol Genet. 2014. PMID: 24030547 Free PMC article.
-
Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development.Clin Genet. 2007 Oct;72(4):296-304. doi: 10.1111/j.1399-0004.2007.00888.x. Clin Genet. 2007. PMID: 17850624 Review.
Cited by
-
Bridging drug discovery through hierarchical subtractive genomics against asd, trpG, and secY of pneumonia causing MDR Staphylococcus aureus.Mol Genet Genomics. 2024 Mar 13;299(1):34. doi: 10.1007/s00438-024-02115-8. Mol Genet Genomics. 2024. PMID: 38478130
-
Computational analysis of the functional and structural impact of the most deleterious missense mutations in the human Protein C.PLoS One. 2023 Nov 28;18(11):e0294417. doi: 10.1371/journal.pone.0294417. eCollection 2023. PLoS One. 2023. PMID: 38015884 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

