A Comparative Analysis of Weizmannia coagulans Genomes Unravels the Genetic Potential for Biotechnological Applications
- PMID: 35328559
- PMCID: PMC8954581
- DOI: 10.3390/ijms23063135
A Comparative Analysis of Weizmannia coagulans Genomes Unravels the Genetic Potential for Biotechnological Applications
Abstract
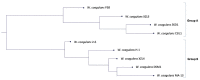
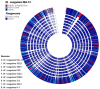
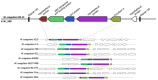
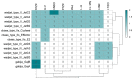
The production of biochemicals requires the use of microbial strains with efficient substrate conversion and excellent environmental robustness, such as Weizmannia coagulans species. So far, the genomes of 47 strains have been sequenced. Herein, we report a comparative genomic analysis of nine strains on the full repertoire of Carbohydrate-Active enZymes (CAZymes), secretion systems, and resistance mechanisms to environmental challenges. Moreover, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) immune system along with CRISPR-associated (Cas) genes, was also analyzed. Overall, this study expands our understanding of the strain's genomic diversity of W. coagulans to fully exploit its potential in biotechnological applications.
Keywords: B. coagulans; CAZymes; CRISPR-Cas systems; bacteriocins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






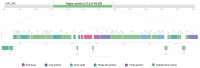
Similar articles
-
Survey of clustered regularly interspaced short palindromic repeats and their associated Cas proteins (CRISPR/Cas) systems in multiple sequenced strains of Klebsiella pneumoniae.BMC Res Notes. 2015 Aug 4;8:332. doi: 10.1186/s13104-015-1285-7. BMC Res Notes. 2015. PMID: 26238567 Free PMC article.
-
Comparative Genomics Analysis of Lactobacillus ruminis from Different Niches.Genes (Basel). 2020 Jan 8;11(1):70. doi: 10.3390/genes11010070. Genes (Basel). 2020. PMID: 31936280 Free PMC article.
-
Occurrence and activity of a type II CRISPR-Cas system in Lactobacillus gasseri.Microbiology (Reading). 2015 Sep;161(9):1752-1761. doi: 10.1099/mic.0.000129. Epub 2015 Jul 9. Microbiology (Reading). 2015. PMID: 26297561
-
CRISPR-Cas system: a powerful tool for genome engineering.Plant Mol Biol. 2014 Jun;85(3):209-18. doi: 10.1007/s11103-014-0188-7. Epub 2014 Mar 18. Plant Mol Biol. 2014. PMID: 24639266 Review.
-
Harnessing CRISPR-Cas systems for bacterial genome editing.Trends Microbiol. 2015 Apr;23(4):225-32. doi: 10.1016/j.tim.2015.01.008. Epub 2015 Feb 17. Trends Microbiol. 2015. PMID: 25698413 Review.
Cited by
-
Whole genome sequencing of the multidrug-resistant Chryseobacterium indologenes isolated from a patient in Brazil.Front Med (Lausanne). 2022 Jul 28;9:931379. doi: 10.3389/fmed.2022.931379. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35966843 Free PMC article.
-
Thermophilic biocatalysts for one-step conversion of citrus waste into lactic acid.Appl Microbiol Biotechnol. 2024 Jan 20;108(1):155. doi: 10.1007/s00253-023-12904-7. Appl Microbiol Biotechnol. 2024. PMID: 38244047 Free PMC article.
-
Comparative genomics reveals carbohydrate enzymatic fluctuations and herbivorous adaptations in arthropods.Comput Struct Biotechnol J. 2024 Oct 18;23:3744-3758. doi: 10.1016/j.csbj.2024.10.027. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39525084 Free PMC article.
-
Metagenomic Analysis of Microbial Community Associated with Food Waste Composting.Appl Biochem Biotechnol. 2025 May;197(5):3503-3520. doi: 10.1007/s12010-025-05203-6. Epub 2025 Feb 17. Appl Biochem Biotechnol. 2025. PMID: 39961944
-
Insight into CAZymes of Alicyclobacillus mali FL18: Characterization of a New Multifunctional GH9 Enzyme.Int J Mol Sci. 2022 Dec 23;24(1):243. doi: 10.3390/ijms24010243. Int J Mol Sci. 2022. PMID: 36613686 Free PMC article.
References
-
- Gupta R.S., Patel S., Saini N., Chen S. Robust demarcation of 17 distinct bacillus species clades, proposed as novel bacillaceae genera, by phylogenomics and comparative genomic analyses: Description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus bacillus limiting it o. Int. J. Syst. Evol. Microbiol. 2020;70:5753–5798. doi: 10.1099/ijsem.0.004475. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

