The Hematopoietic TALE-Code Shows Normal Activity of IRX1 in Myeloid Progenitors and Reveals Ectopic Expression of IRX3 and IRX5 in Acute Myeloid Leukemia
- PMID: 35328612
- PMCID: PMC8952210
- DOI: 10.3390/ijms23063192
The Hematopoietic TALE-Code Shows Normal Activity of IRX1 in Myeloid Progenitors and Reveals Ectopic Expression of IRX3 and IRX5 in Acute Myeloid Leukemia
Abstract
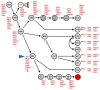
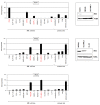
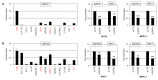
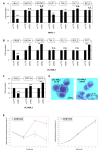
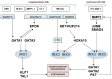
Homeobox genes encode transcription factors that control basic developmental decisions. Knowledge of their hematopoietic activities casts light on normal and malignant immune cell development. Recently, we constructed the so-called lymphoid TALE-code that codifies expression patterns of all active TALE class homeobox genes in early hematopoiesis and lymphopoiesis. Here, we present the corresponding myeloid TALE-code to extend this gene signature, covering the entire hematopoietic system. The collective data showed expression patterns for eleven TALE homeobox genes and highlighted the exclusive expression of IRX1 in megakaryocyte-erythroid progenitors (MEPs), implicating this TALE class member in a specific myeloid differentiation process. Analysis of public profiling data from acute myeloid leukemia (AML) patients revealed aberrant activity of IRX1 in addition to IRX3 and IRX5, indicating an oncogenic role for these TALE homeobox genes when deregulated. Screening of RNA-seq data from 100 leukemia/lymphoma cell lines showed overexpression of IRX1, IRX3, and IRX5 in megakaryoblastic and myelomonocytic AML cell lines, chosen as suitable models for studying the regulation and function of these homeo-oncogenes. Genomic copy number analysis of IRX-positive cell lines demonstrated chromosomal amplification of the neighboring IRX3 and IRX5 genes at position 16q12 in MEGAL, underlying their overexpression in this cell line model. Comparative gene expression analysis of these cell lines revealed candidate upstream factors and target genes, namely the co-expression of GATA1 and GATA2 together with IRX1, and of BMP2 and HOXA10 with IRX3/IRX5. Subsequent knockdown and stimulation experiments in AML cell lines confirmed their activating impact in the corresponding IRX gene expression. Furthermore, we demonstrated that IRX1 activated KLF1 and TAL1, while IRX3 inhibited GATA1, GATA2, and FST. Accordingly, we propose that these regulatory relationships may represent major physiological and oncogenic activities of IRX factors in normal and malignant myeloid differentiation, respectively. Finally, the established myeloid TALE-code is a useful tool for evaluating TALE homeobox gene activities in AML.
Keywords: HOX-code; NKL-code; PBX1; TALE-code; TBX-code; homeodomain.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wilson N.K., Foster S.D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E., et al. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

