DkmiR397 Regulates Proanthocyanidin Biosynthesis via Negative Modulating DkLAC2 in Chinese PCNA Persimmon
- PMID: 35328620
- PMCID: PMC8951489
- DOI: 10.3390/ijms23063200
DkmiR397 Regulates Proanthocyanidin Biosynthesis via Negative Modulating DkLAC2 in Chinese PCNA Persimmon
Abstract
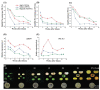
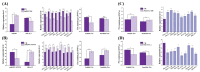
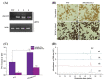
Persimmon fruits accumulate a large amount of proanthocyanidins (PAs), which makes an astringent sensation. Proanthocyanidins (PAs) are the polymers of flavan-3-ols stored in plant vacuoles under laccase activation. A laccase gene, DkLAC2, is putatively involved in PAs biosynthesis and regulated by microRNA (DkmiR397) in persimmon. However, the polymerization of PAs in association with miRNA397 still needs to be explored in persimmon. Here, we identified pre-DkmiR397 and its target gene DkLAC2 in 'Eshi 1' persimmon. Histochemical staining with GUS and dual luciferase assay both confirmed DkmiR397-DkLAC2 binding after co-transformation in tobacco leaves. Diverse expression patterns of DkLAC2 and DkmiR397 were exhibited during persimmon fruit development stages. Moreover, a contrasting expression pattern was also observed after the combined DkLAC2-miR397 transformation in persimmon leaves, suggesting that DkmiR397 might be a negative regulator of DkLAC2. Similarly, the transient transformation of DkmiR397 in persimmon fruit discs in vitro also reduced PA accumulation by repressing DkLAC2, whereas the up-regulation of DkLAC2 increased the accumulation of PAs by short tandem target mimic STTM-miR397. A similar expression pattern was observed when overexpressing of DkLAC2 in Arabidopsis wild type (WT) and overexpression of DkLAC2, DkmiR397 in persimmon leaf callus. Our results revealed that the role of DkmiR397 repressed the expression of DkLAC2 concerning PA biosynthesis, providing a potential target for the manipulation of PAs metabolism in persimmon.
Keywords: laccase; microRNA; persimmon; polymerization; tannin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Molecular cloning and functional characterization of DkMATE1 involved in proanthocyanidin precursor transport in persimmon (Diospyros kaki Thunb.) fruit.Plant Physiol Biochem. 2016 Nov;108:241-250. doi: 10.1016/j.plaphy.2016.07.016. Epub 2016 Jul 19. Plant Physiol Biochem. 2016. PMID: 27472890
-
Isolation and characterization of a laccase gene potentially involved in proanthocyanidin polymerization in Oriental persimmon (Diospyros kaki Thunb.) fruit.Mol Biol Rep. 2013 Apr;40(4):2809-20. doi: 10.1007/s11033-012-2296-2. Epub 2012 Dec 9. Mol Biol Rep. 2013. PMID: 23224657
-
DkDTX1/MATE1 mediates the accumulation of proanthocyanidin and affects astringency in persimmon.Plant Cell Environ. 2024 Dec;47(12):5205-5219. doi: 10.1111/pce.15092. Epub 2024 Aug 22. Plant Cell Environ. 2024. PMID: 39169830
-
Molecular Controls of Proanthocyanidin Synthesis and Structure: Prospects for Genetic Engineering in Crop Plants.J Agric Food Chem. 2018 Sep 26;66(38):9882-9888. doi: 10.1021/acs.jafc.8b02950. Epub 2018 Sep 13. J Agric Food Chem. 2018. PMID: 30139248 Review.
-
Activity and potential mechanisms of action of persimmon tannins according to their structures: A review.Int J Biol Macromol. 2023 Jul 1;242(Pt 3):125120. doi: 10.1016/j.ijbiomac.2023.125120. Epub 2023 May 30. Int J Biol Macromol. 2023. PMID: 37263329 Review.
Cited by
-
Responses of sorghum to cold stress: A review focused on molecular breeding.Front Plant Sci. 2023 Feb 23;14:1124335. doi: 10.3389/fpls.2023.1124335. eCollection 2023. Front Plant Sci. 2023. PMID: 36909409 Free PMC article. Review.
-
Research on multi-cluster green persimmon detection method based on improved Faster RCNN.Front Plant Sci. 2023 Jun 6;14:1177114. doi: 10.3389/fpls.2023.1177114. eCollection 2023. Front Plant Sci. 2023. PMID: 37346117 Free PMC article.
-
Analysis of Homologous Regions of Small RNAs MIR397 and MIR408 Reveals the Conservation of Microsynteny among Rice Crop-Wild Relatives.Cells. 2022 Nov 2;11(21):3461. doi: 10.3390/cells11213461. Cells. 2022. PMID: 36359857 Free PMC article.
-
Regulatory mechanisms and metabolic changes of miRNA during leaf color change in the bud mutation branches of Acer pictum subsp. mono.Front Plant Sci. 2023 Jan 12;13:1047452. doi: 10.3389/fpls.2022.1047452. eCollection 2022. Front Plant Sci. 2023. PMID: 36714704 Free PMC article.
-
Overview of Repressive miRNA Regulation by Short Tandem Target Mimic (STTM): Applications and Impact on Plant Biology.Plants (Basel). 2023 Feb 3;12(3):669. doi: 10.3390/plants12030669. Plants (Basel). 2023. PMID: 36771753 Free PMC article. Review.
References
-
- Luo Z., Wang R. Persimmon in China: Domestication and traditional utilizations of genetic resources. Adv. Hortic. Sci. 2008;22:239–243. doi: 10.1400/100648. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

