Inducible Systemic Gcn1 Deletion in Mice Leads to Transient Body Weight Loss upon Tamoxifen Treatment Associated with Decrease of Fat and Liver Glycogen Storage
- PMID: 35328622
- PMCID: PMC8949040
- DOI: 10.3390/ijms23063201
Inducible Systemic Gcn1 Deletion in Mice Leads to Transient Body Weight Loss upon Tamoxifen Treatment Associated with Decrease of Fat and Liver Glycogen Storage
Abstract
GCN1 is an evolutionarily-conserved ribosome-binding protein that mediates the amino acid starvation response as well as the ribotoxic stress response. We previously demonstrated that Gcn1 mutant mice lacking the GCN2-binding domain suffer from growth retardation and postnatal lethality via GCN2-independent mechanisms, while Gcn1-null mice die early in embryonic development. In this study, we explored the role of GCN1 in adult mice by generating tamoxifen-inducible conditional knockout (CKO) mice. Unexpectedly, the Gcn1 CKO mice showed body weight loss during tamoxifen treatment, which gradually recovered following its cessation. They also showed decreases in liver weight, hepatic glycogen and lipid contents, blood glucose and non-esterified fatty acids, and visceral white adipose tissue weight with no changes in food intake and viability. A decrease of serum VLDL suggested that hepatic lipid supply to the peripheral tissues was primarily impaired. Liver proteomic analysis revealed the downregulation of mitochondrial β-oxidation that accompanied increases of peroxisomal β-oxidation and aerobic glucose catabolism that maintain ATP levels. These findings show the involvement of GCN1 in hepatic lipid metabolism during tamoxifen treatment in adult mice.
Keywords: GCN1; VLDL; body weight loss; lipid storage; tamoxifen toxicity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
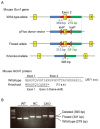

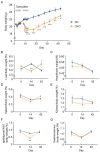
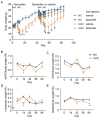
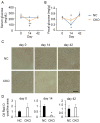
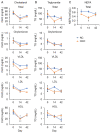
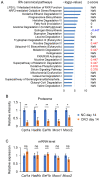

References
-
- Yamazaki H., Kasai S., Mimura J., Ye P., Inose-Maruyama A., Tanji K., Wakabayashi K., Mizuno S., Sugiyama F., Takahashi S., et al. Ribosome binding protein GCN1 regulates the cell cycle and cell proliferation and is essential for the embryonic development of mice. PLoS Genet. 2020;16:e1008693. doi: 10.1371/journal.pgen.1008693. - DOI - PMC - PubMed
-
- Zhang P., McGrath B.C., Reinert J., Olsen D.S., Lei L., Gill S., Wek S.A., Vattem K.M., Wek R.C., Kimball S.R., et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-6688.2002. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

