Regulatory Mechanism of Transcription Factor AhHsf Modulates AhHsp70 Transcriptional Expression Enhancing Heat Tolerance in Agasicles hygrophila (Coleoptera: Chrysomelidae)
- PMID: 35328631
- PMCID: PMC8955217
- DOI: 10.3390/ijms23063210
Regulatory Mechanism of Transcription Factor AhHsf Modulates AhHsp70 Transcriptional Expression Enhancing Heat Tolerance in Agasicles hygrophila (Coleoptera: Chrysomelidae)
Abstract
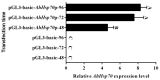
Agasicles hygrophila is a classical biological agent used to control alligator weed (Alternanthera philoxeroides). Previous research has indicated that the heat shock factor (HSF) is involved in regulating the transcriptional expression of Hsp70 in response to heat resistance in A. hygrophila. However, the regulatory mechanism by which AhHsf regulates the expression of AhHsp70 remains largely unknown. Here, we identified and cloned a 944 bp AhHsp70 promoter (AhHsp70p) region from A. hygrophila. Subsequent bioinformatics analysis revealed that the AhHsp70p sequence contains multiple functional elements and has a common TATA box approximately 30 bp upstream of the transcription start site, with transcription commencing at a purine base approximately 137 bp upstream of ATG. Promoter deletion analyses revealed that the sequence from -944 to -744 bp was the core regulatory region. A dual-luciferase reporter assay indicated that overexpressed AhHsf significantly enhanced the activity of AhHsp70p. Furthermore, qPCR showed that AhHsp70 expression increased with time in Spodoptera frugiperda (Sf9) cells, and AhHsf overexpression significantly upregulated AhHsp70 expression in vitro. Characterization of the upstream regulatory mechanisms demonstrated that AhHsf binds to upstream cis-acting elements in the promoter region of AhHsp70 from -944 to -744 bp to activate the AhHSF-AhHSP pathway at the transcriptional level to protect A. hygrophila from high temperature damage. Furthermore, we proposed a molecular model of AhHsf modulation of AhHsp70 transcription following heat shock in A. hygrophila. The findings of this study suggest that enhancing the heat tolerance of A. hygrophila by modulating the upstream pathways of the Hsp family can improve the biocontrol of A. philoxeroides.
Keywords: Agasicles hygrophila; cell transfection; heat shock protein 70 promoter (Hsp70p); inverse PCR (I-PCR); real-time quantitative PCR (RT-qPCR); transcription factor AhHsf.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures





References
-
- Bale J.S., Masters G.J., Hodkinson I.D., Awmack C., Bezemer T.M., Brown V.K., Butterfield J., Buse A., Coulson J.C., Farrar J., et al. Herbivory in global climate change research: Direct effects of rising temperature on insect herbivores. Glob. Chang. Biol. 2002;8:1–16. doi: 10.1046/j.1365-2486.2002.00451.x. - DOI
-
- Yu H., Wan F.H., Guo J.Y. cDNA Cloning of Heat Shock Protein Genes and Their Expression in an Indigenous Cryptic Species of the Whitefly Bemisia tabaci Complex from China. J. Integr. Agric. 2012;11:293–302. doi: 10.1016/S2095-3119(12)60013-6. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

