Insights in Post-Translational Modifications: Ubiquitin and SUMO
- PMID: 35328702
- PMCID: PMC8952880
- DOI: 10.3390/ijms23063281
Insights in Post-Translational Modifications: Ubiquitin and SUMO
Abstract
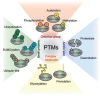
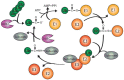
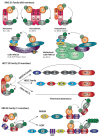
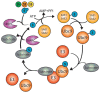
Both ubiquitination and SUMOylation are dynamic post-translational modifications that regulate thousands of target proteins to control virtually every cellular process. Unfortunately, the detailed mechanisms of how all these cellular processes are regulated by both modifications remain unclear. Target proteins can be modified by one or several moieties, giving rise to polymers of different morphology. The conjugation cascades of both modifications comprise a few activating and conjugating enzymes but close to thousands of ligating enzymes (E3s) in the case of ubiquitination. As a result, these E3s give substrate specificity and can form polymers on a target protein. Polymers can be quickly modified forming branches or cleaving chains leading the target protein to its cellular fate. The recent development of mass spectrometry(MS) -based approaches has increased the understanding of ubiquitination and SUMOylation by finding essential modified targets in particular signaling pathways. Here, we perform a concise overview comprising from the basic mechanisms of both ubiquitination and SUMOylation to recent MS-based approaches aimed to find specific targets for particular E3 enzymes.
Keywords: E3 enzymes; SUMO; proteomics; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

