The Human Induced Pluripotent Stem Cell Test as an Alternative Method for Embryotoxicity Testing
- PMID: 35328717
- PMCID: PMC8950674
- DOI: 10.3390/ijms23063295
The Human Induced Pluripotent Stem Cell Test as an Alternative Method for Embryotoxicity Testing
Abstract
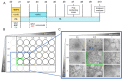
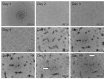
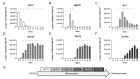
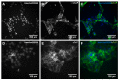
The evaluation of substances for their potency to induce embryotoxicity is controlled by safety regulations. Test guidelines for reproductive and developmental toxicity rely mainly on animal studies, which make up the majority of animal usage in regulatory toxicology. Therefore, there is an urgent need for alternative in vitro methods to follow the 3R principles. To improve human safety, cell models based on human cells are of great interest to overcome species differences. Here, human induced pluripotent stem cells (hiPSCs) are an ideal cell source as they largely recapitulate embryonic stem cells without bearing ethical concerns and they are able to differentiate into most cell types of the human body. Here, we set up and characterized a fetal bovine serum (FBS)-free hiPSC-based in vitro test method, called the human induced pluripotent stem cell test (hiPS Test), to evaluate the embryotoxic potential of substances. After 10 days in culture, hiPSCs develop into beating cardiomyocytes. As terminal endpoint evaluations, cell viability, qPCR analyses as well as beating frequency and area of beating cardiomyocytes by video analyses are measured. The embryotoxic positive and non-embryotoxic negative controls, 5-Fluorouracil (5-FU) and Penicillin G (PenG), respectively, were correctly assessed in the hiPS Test. More compounds need to be screened in the future for defining the assay's applicability domain, which will inform us of the suitability of the hiPS Test for detecting adverse effects of substances on embryonic development.
Keywords: CardioVision; cardiomyocytes; developmental toxicity; embryotoxicity; hiPS Test; hiPSC; in vitro; video analyses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
An in vitro embryotoxicity assay based on the disturbance of the differentiation of murine embryonic stem cells into endothelial cells. II. Testing of compounds.Toxicol In Vitro. 2007 Dec;21(8):1631-40. doi: 10.1016/j.tiv.2007.06.014. Epub 2007 Jul 18. Toxicol In Vitro. 2007. PMID: 17719739
-
An Evaluation of Human Induced Pluripotent Stem Cells to Test for Cardiac Developmental Toxicity.Int J Mol Sci. 2021 Jul 29;22(15):8114. doi: 10.3390/ijms22158114. Int J Mol Sci. 2021. PMID: 34360880 Free PMC article.
-
Improvement of the embryonic stem cell test endpoint analysis by use of field potential detection.J Toxicol Sci. 2010 Oct;35(5):619-29. doi: 10.2131/jts.35.619. J Toxicol Sci. 2010. PMID: 20930457
-
Embryonic stem cell test: stem cell use in predicting developmental cardiotoxicity and osteotoxicity.Methods Mol Biol. 2012;889:147-79. doi: 10.1007/978-1-61779-867-2_10. Methods Mol Biol. 2012. PMID: 22669664 Review.
-
Embryonic stem cells as an in vitro model for mutagenicity, cytotoxicity and embryotoxicity studies: present state and future prospects.Toxicol In Vitro. 2001 Dec;15(6):741-53. doi: 10.1016/s0887-2333(01)00074-1. Toxicol In Vitro. 2001. PMID: 11698176 Review.
Cited by
-
Investigating the applicability domain of the hiPSC-based PluriLum assay: an embryotoxicity assessment of chemicals and drugs.Arch Toxicol. 2024 Apr;98(4):1209-1224. doi: 10.1007/s00204-023-03675-1. Epub 2024 Feb 4. Arch Toxicol. 2024. PMID: 38311648 Free PMC article.
-
Molecular changes in endometrium origin stromal cells during initiation of cardiomyogenic differentiation induced with Decitabine, Angiotensin II and TGF- β1.Sci Rep. 2024 Jul 23;14(1):16966. doi: 10.1038/s41598-024-68108-0. Sci Rep. 2024. PMID: 39043870 Free PMC article.
-
Developmental Toxicity Studies: The Path towards Humanized 3D Stem Cell-Based Models.Int J Mol Sci. 2023 Mar 2;24(5):4857. doi: 10.3390/ijms24054857. Int J Mol Sci. 2023. PMID: 36902285 Free PMC article. Review.
-
The Mechanism of Houttuynia cordata Embryotoxicity Was Explored in Combination with an Experimental Model and Network Pharmacology.Toxins (Basel). 2023 Jan 13;15(1):73. doi: 10.3390/toxins15010073. Toxins (Basel). 2023. PMID: 36668893 Free PMC article.
-
Exposure to Toxicants Affects Everyone, Especially the Very Young.Int J Mol Sci. 2022 Jun 29;23(13):7232. doi: 10.3390/ijms23137232. Int J Mol Sci. 2022. PMID: 35806235 Free PMC article.
References
-
- Miller B.F., Keane C., O’Toole M.T. Encyclopedia & Dictionary of Medicine, Nursing & Allied Health. 7th ed. W B Saunders Co; Philadelphia, PA, USA: 2003.
-
- MacDorman M.F., Gregory E.C.W. Fetal and Perinatal Mortality: United States, 2013. Natl. Vital Stat. Rep. 2015;64:24. - PubMed
-
- DeSesso J.M. Future of Developmental Toxicity Testing. Curr. Opin. Toxicol. 2017;3:1–5. doi: 10.1016/j.cotox.2017.04.001. - DOI
-
- Weinhold B. Environmental Factors in Birth Defects: What We Need to Know. Environ. Health Perspect. 2009;117:A440–A447. doi: 10.1289/ehp.117-a440. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

