A White Plaque, Associated with Genomic Deletion, Derived from M13KE-Based Peptide Library Is Enriched in a Target-Unrelated Manner during Phage Display Biopanning Due to Propagation Advantage
- PMID: 35328728
- PMCID: PMC8950111
- DOI: 10.3390/ijms23063308
A White Plaque, Associated with Genomic Deletion, Derived from M13KE-Based Peptide Library Is Enriched in a Target-Unrelated Manner during Phage Display Biopanning Due to Propagation Advantage
Abstract
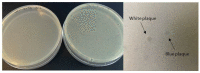
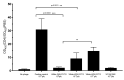
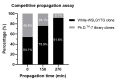
The nonspecific enrichment of target-unrelated peptides during biopanning remains a major drawback for phage display technology. The commercial Ph.D.TM-7 phage display library is used extensively for peptide discovery. This library is based on the M13KE vector, which carries the lacZα sequence, leading to the formation of blue plaques on IPTG-X-gal agar plates. In the current study, we report the isolation of a fast-propagating white clone (displaying WSLGYTG peptide) identified through screening against a recombinant protein. Sanger sequencing demonstrated that white plaques are not contamination from environmental M13-like phages, but derive from the library itself. Whole genome sequencing revealed that the white color of the plaques results from a large 827-nucleotide genomic deletion. The phenotypic characterization of propagation capacity through plaque count- and NGS-based competitive propagation assay supported the higher propagation rate of Ph-WSLGYTG clone compared with the library. According to our data, white plaques are likely to arise endogenously in Ph.D. libraries due to mutations in the M13KE genome and should not always be viewed as exogenous contamination. Our findings also led to the conclusion that the deletion observed here might be an ancestral mutation already present in the naïve library, which causes target-unrelated nonspecific enrichment of white clone during biopanning due to propagation advantage.
Keywords: M13KE; Ph.D.TM-7 peptide library; biopanning; competitive propagation; genomic deletion; lacZα sequence; phage display; propagation-related TUP; white plaque.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Propagation Capacity of Phage Display Peptide Libraries Is Affected by the Length and Conformation of Displayed Peptide.Molecules. 2023 Jul 10;28(14):5318. doi: 10.3390/molecules28145318. Molecules. 2023. PMID: 37513190 Free PMC article.
-
Identification and characterization of mutant clones with enhanced propagation rates from phage-displayed peptide libraries.Anal Biochem. 2014 Oct 1;462:35-43. doi: 10.1016/j.ab.2014.06.007. Epub 2014 Jun 19. Anal Biochem. 2014. PMID: 24952360
-
Phage display biopanning and isolation of target-unrelated peptides: in search of nonspecific binders hidden in a combinatorial library.Amino Acids. 2016 Dec;48(12):2699-2716. doi: 10.1007/s00726-016-2329-6. Epub 2016 Sep 20. Amino Acids. 2016. PMID: 27650972 Review.
-
Using NGS to Uncover the Corruption of a Peptide Phage Display Selection.Curr Issues Mol Biol. 2024 Sep 21;46(9):10590-10605. doi: 10.3390/cimb46090627. Curr Issues Mol Biol. 2024. PMID: 39329979 Free PMC article.
-
Biased selection of propagation-related TUPs from phage display peptide libraries.Amino Acids. 2017 Aug;49(8):1293-1308. doi: 10.1007/s00726-017-2452-z. Epub 2017 Jun 29. Amino Acids. 2017. PMID: 28664268 Review.
Cited by
-
A comparative analysis of sequence composition in different lots of a phage display peptide library during amplification.Virol J. 2025 Feb 1;22(1):24. doi: 10.1186/s12985-024-02600-x. Virol J. 2025. PMID: 39893369 Free PMC article.
-
Depth of Sequencing Plays a Determining Role in the Characterization of Phage Display Peptide Libraries by NGS.Int J Mol Sci. 2023 Mar 11;24(6):5396. doi: 10.3390/ijms24065396. Int J Mol Sci. 2023. PMID: 36982469 Free PMC article.
-
On the origin of non-specific binders isolated in the selection of phage display peptide libraries.Front Microbiol. 2025 Jun 4;16:1571679. doi: 10.3389/fmicb.2025.1571679. eCollection 2025. Front Microbiol. 2025. PMID: 40535010 Free PMC article. Review.
-
Analysis of Compositional Bias in a Commercial Phage Display Peptide Library by Next-Generation Sequencing.Viruses. 2022 Oct 29;14(11):2402. doi: 10.3390/v14112402. Viruses. 2022. PMID: 36366500 Free PMC article.
-
Propagation Capacity of Phage Display Peptide Libraries Is Affected by the Length and Conformation of Displayed Peptide.Molecules. 2023 Jul 10;28(14):5318. doi: 10.3390/molecules28145318. Molecules. 2023. PMID: 37513190 Free PMC article.
References
-
- Instruction Manual Ph.D. Phage Display Libraries. no. 5.0. 2020. [(accessed on 20 January 2022)]. Available online: https://international.neb.com/-/media/nebus/files/manuals/manuale8100_e8....
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

