Pericentromeric Non-Coding DNA Transcription Is Associated with Niche Impairment in Patients with Ineffective or Partially Effective Multiple Myeloma Treatment
- PMID: 35328779
- PMCID: PMC8951104
- DOI: 10.3390/ijms23063359
Pericentromeric Non-Coding DNA Transcription Is Associated with Niche Impairment in Patients with Ineffective or Partially Effective Multiple Myeloma Treatment
Abstract
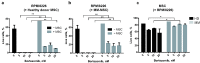
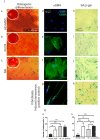
Mesenchymal stromal cells (MSC) 'educated' by tumor cells are an essential component of the multiple myeloma (MM) tumor microenvironment (TME) involved in tumor progression. Transcription of tandemly repeated (TR) non-coding DNA is often activated in many tumors and is required for tumor progression and cancer cells genome reorganization. The aim of the work was to study functional properties including the TR DNA transcription profile of MSC from the hematopoietic niche of treated MM patients. Healthy donors (HD) and patients after bortezomib-based treatment (with partial or complete response, PoCR, and non-responders, NR) were enrolled in the study. Their trephine biopsies were examined histologically to evaluate the hematopoietic niche. MSC cultures obtained from the biopsies were used for evaluation of the proliferation rate, osteogenic differentiation, presence of tumor MSC markers, resistance to bortezomib, and pericentromeric TR DNA transcription level. The MSC 'education' by multiple myeloma cells was mimicked in co-culture experiments with or without bortezomib. The TR DNA transcription profile was accessed. The histological examination revealed the persistence of the tumor microenvironment (especially of the vasculature) in treated patients. In co-culture experiments, MSC of bortezomib-treated patients were more resistant to bortezomib and protected cancer MM cells of the RPMI8226 cell line more effectively than HD-MSC did. The MSC obtained from PoCR and NR samples differed in their functional properties (proliferation capacity, osteogenic potential, and cancer-associated fibroblasts markers). Transcriptome analysis revealed activation of the TR transcription in cells of non-hematopoietic origin from NR patients' bone marrow. The pericentromeric TR DNA of HS2/HS3 families was among the most upregulated in stromal MSC but not in cancer cells. The highest level of transcription was observed in NR-MSC. Transcription of HS2/HS3 was not detected in healthy donors MSC unless they were co-cultured with MM cancer cells and acquired cancer-associated phenotype. Treatment with TNFα downregulated HS2/HS3 transcription in MSC and upregulated in MM cells. Our results suggest that the hematopoietic niche retains the cancer-associated phenotype after treatment. Pericentromeric non-coding DNA transcription is associated with the MSC cancer-associated phenotype in patients with ineffective or partially effective multiple myeloma treatment.
Keywords: human satellite 2; human satellite 3; long non-coding RNA; mesenchymal stromal cells; microvessels; multiple myeloma; non-coding DNA transcripts; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Bessmeltsev S.S. Multiple myeloma (pathogenesis, clinical features, diagnosis, differential diagnosis). Part I. Clin. Oncohematol. 2013;6:237–257.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

