The Pathogen-Induced MATE Gene TaPIMA1 Is Required for Defense Responses to Rhizoctonia cerealis in Wheat
- PMID: 35328796
- PMCID: PMC8950252
- DOI: 10.3390/ijms23063377
The Pathogen-Induced MATE Gene TaPIMA1 Is Required for Defense Responses to Rhizoctonia cerealis in Wheat
Abstract
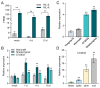
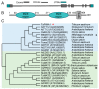
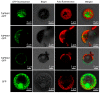
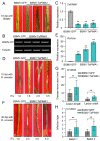
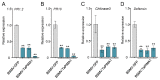
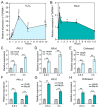
The sharp eyespot, mainly caused by the soil-borne fungus Rhizoctonia cerealis, is a devastating disease endangering production of wheat (Triticum aestivum). Multi-Antimicrobial Extrusion (MATE) family genes are widely distributed in plant species, but little is known about MATE functions in wheat disease resistance. In this study, we identified TaPIMA1, a pathogen-induced MATE gene in wheat, from RNA-seq data. TaPIMA1 expression was induced by Rhizoctonia cerealis and was higher in sharp eyespot-resistant wheat genotypes than in susceptible wheat genotypes. Molecular biology assays showed that TaPIMA1 belonged to the MATE family, and the expressed protein could distribute in the cytoplasm and plasma membrane. Virus-Induced Gene Silencing plus disease assessment indicated that knock-down of TaPIMA1 impaired resistance of wheat to sharp eyespot and down-regulated the expression of defense genes (Defensin, PR10, PR1.2, and Chitinase3). Furthermore, TaPIMA1 was rapidly induced by exogenous H2O2 and jasmonate (JA) treatments, which also promoted the expression of pathogenesis-related genes. These results suggested that TaPIMA1 might positively regulate the defense against R. cerealis by up-regulating the expression of defense-associated genes in H2O2 and JA signal pathways. This study sheds light on the role of MATE transporter in wheat defense to Rhizoctonia cerealis and provides a potential gene for improving wheat resistance against sharp eyespot.
Keywords: Rhizoctonia cerealis; TaPIMA1; defense; multi-antimicrobial extrusion family; wheat (Triticum aestivum).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes.Sci Rep. 2016 Jul 1;6:28777. doi: 10.1038/srep28777. Sci Rep. 2016. PMID: 27364458 Free PMC article.
-
The cysteine-rich receptor-like kinase TaCRK3 contributes to defense against Rhizoctonia cerealis in wheat.J Exp Bot. 2021 Oct 26;72(20):6904-6919. doi: 10.1093/jxb/erab328. J Exp Bot. 2021. PMID: 34254642
-
The wheat AGC kinase TaAGC1 is a positive contributor to host resistance to the necrotrophic pathogen Rhizoctonia cerealis.J Exp Bot. 2015 Nov;66(21):6591-603. doi: 10.1093/jxb/erv367. Epub 2015 Jul 27. J Exp Bot. 2015. PMID: 26220083 Free PMC article.
-
The wheat NB-LRR gene TaRCR1 is required for host defence response to the necrotrophic fungal pathogen Rhizoctonia cerealis.Plant Biotechnol J. 2017 Jun;15(6):674-687. doi: 10.1111/pbi.12665. Epub 2017 Mar 1. Plant Biotechnol J. 2017. PMID: 27862842 Free PMC article.
-
The escalating threat of Rhizoctonia cerealis, the causal agent of sharp eyespot in wheat.Pest Manag Sci. 2011 Nov;67(11):1411-9. doi: 10.1002/ps.2236. Epub 2011 Jul 1. Pest Manag Sci. 2011. PMID: 21726039 Review.
Cited by
-
Multidrug and Toxic Compound Extrusion Transporters: Ubiquitous Multifaceted Proteins in Microbes, Plants, and Their Interactions.Microorganisms. 2024 Nov 27;12(12):2433. doi: 10.3390/microorganisms12122433. Microorganisms. 2024. PMID: 39770636 Free PMC article. Review.
-
Pathogen lifestyle determines host genetic signature of quantitative disease resistance loci in oilseed rape (Brassica napus).Theor Appl Genet. 2024 Mar 2;137(3):65. doi: 10.1007/s00122-024-04569-1. Theor Appl Genet. 2024. PMID: 38430276 Free PMC article.
-
Comprehensive Identification and Expression Analysis of the Multidrug and Toxic Compound Extrusion (MATE) Gene Family in Brachypodium distachyon.Plants (Basel). 2024 Sep 15;13(18):2586. doi: 10.3390/plants13182586. Plants (Basel). 2024. PMID: 39339561 Free PMC article.
-
Overexpression of TaMYC2 confers freeze tolerance by ICE-CBF-COR module in Arabidopsis thaliana.Front Plant Sci. 2022 Nov 14;13:1042889. doi: 10.3389/fpls.2022.1042889. eCollection 2022. Front Plant Sci. 2022. PMID: 36466238 Free PMC article.
References
-
- Hoeven E., Bollen G.J. Effect of benomyl on soil fungi associated with rye. 1. Effect on the incidence of sharp eyespot caused by Rhizoctonia cerealis. Neth. J. Plant Pathol. 1980;86:163–180. doi: 10.1007/BF01989709. - DOI
-
- Murray D., Burpee L.L. Ceratobasidium cereale sp.nov., the teleomorph of Rhizoctonia cerealis. Trans. Brit. Mycol. Soc. 1984;82:170–172. doi: 10.1016/S0007-1536(84)80227-2. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

