Improved Binding Affinity of Omicron's Spike Protein for the Human Angiotensin-Converting Enzyme 2 Receptor Is the Key behind Its Increased Virulence
- PMID: 35328828
- PMCID: PMC8955673
- DOI: 10.3390/ijms23063409
Improved Binding Affinity of Omicron's Spike Protein for the Human Angiotensin-Converting Enzyme 2 Receptor Is the Key behind Its Increased Virulence
Abstract
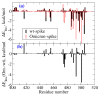
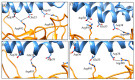
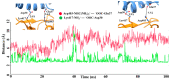
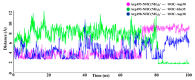
The new variant of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), Omicron, has been quickly spreading in many countries worldwide. Compared to the original virus, Omicron is characterized by several mutations in its genomic region, including the spike protein's receptor-binding domain (RBD). We have computationally investigated the interaction between the RBD of both the wild type and Omicron variant of SARS-CoV-2 with the human angiotensin-converting enzyme 2 (hACE2) receptor using molecular dynamics and molecular mechanics-generalized Born surface area (MM-GBSA)-based binding free energy calculations. The mode of the interaction between Omicron's RBD with the hACE2 receptor is similar to the original SARS-CoV-2 RBD except for a few key differences. The binding free energy difference shows that the spike protein of Omicron has an increased affinity for the hACE2 receptor. The mutated residues in the RBD showed strong interactions with a few amino acid residues of hACE2. More specifically, strong electrostatic interactions (salt bridges) and hydrogen bonding were observed between R493 and R498 residues of the Omicron RBD with D30/E35 and D38 residues of the hACE2, respectively. Other mutated amino acids in the Omicron RBD, e.g., S496 and H505, also exhibited hydrogen bonding with the hACE2 receptor. A pi-stacking interaction was also observed between tyrosine residues (RBD-Tyr501: hACE2-Tyr41) in the complex, which contributes majorly to the binding free energies and suggests that this is one of the key interactions stabilizing the formation of the complex. The resulting structural insights into the RBD:hACE2 complex, the binding mode information within it, and residue-wise contributions to the free energy provide insight into the increased transmissibility of Omicron and pave the way to design and optimize novel antiviral agents.
Keywords: Omicron; human angiotensin-converting enzyme 2 (hACE2); molecular dynamics simulation; molecular mechanics-generalized Born surface area (MM-GBSA); receptor-binding domain (RBD); receptor-binding motif (RBM); severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment.J Med Virol. 2022 Oct;94(10):4780-4791. doi: 10.1002/jmv.27927. Epub 2022 Jun 16. J Med Virol. 2022. PMID: 35680610 Free PMC article.
-
E484K mutation in SARS-CoV-2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: Binding free energy calculation studies.J Mol Graph Model. 2021 Dec;109:108035. doi: 10.1016/j.jmgm.2021.108035. Epub 2021 Sep 17. J Mol Graph Model. 2021. PMID: 34562851 Free PMC article.
-
Impact of African-Specific ACE2 Polymorphisms on Omicron BA.4/5 RBD Binding and Allosteric Communication Within the ACE2-RBD Protein Complex.Int J Mol Sci. 2025 Feb 6;26(3):1367. doi: 10.3390/ijms26031367. Int J Mol Sci. 2025. PMID: 39941135 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Surface charge changes in spike RBD mutations of SARS-CoV-2 and its variant strains alter the virus evasiveness via HSPGs: A review and mechanistic hypothesis.Front Public Health. 2022 Aug 24;10:952916. doi: 10.3389/fpubh.2022.952916. eCollection 2022. Front Public Health. 2022. PMID: 36091499 Free PMC article. Review.
Cited by
-
The Effect of Select SARS-CoV-2 N-Linked Glycan and Variant of Concern Spike Protein Mutations on C-Type Lectin-Receptor-Mediated Infection.Viruses. 2023 Sep 9;15(9):1901. doi: 10.3390/v15091901. Viruses. 2023. PMID: 37766307 Free PMC article.
-
Atomistic insights into the binding of SARS-CoV-2 spike receptor binding domain with the human ACE2 receptor: The importance of residue 493.J Mol Graph Model. 2023 Jan;118:108360. doi: 10.1016/j.jmgm.2022.108360. Epub 2022 Oct 22. J Mol Graph Model. 2023. PMID: 36401897 Free PMC article.
-
Probing Mechanisms of Binding and Allostery in the SARS-CoV-2 Spike Omicron Variant Complexes with the Host Receptor: Revealing Functional Roles of the Binding Hotspots in Mediating Epistatic Effects and Communication with Allosteric Pockets.Int J Mol Sci. 2022 Sep 29;23(19):11542. doi: 10.3390/ijms231911542. Int J Mol Sci. 2022. PMID: 36232845 Free PMC article.
-
Impact of Missense Mutations on Spike Protein Stability and Binding Affinity in the Omicron Variant.Viruses. 2024 Jul 17;16(7):1150. doi: 10.3390/v16071150. Viruses. 2024. PMID: 39066312 Free PMC article.
-
Omicron Coronavirus: pH-Dependent Electrostatic Potential and Energy of Association of Spike Protein to ACE2 Receptor.Viruses. 2023 Aug 17;15(8):1752. doi: 10.3390/v15081752. Viruses. 2023. PMID: 37632094 Free PMC article.
References
-
- European Centre for Disease Prevention and Control. SARS-CoV-2 Variants of Concern as of 13 January 2022. [(accessed on 10 December 2021)]. Available online: https://www.ecdc.europa.eu/en/covid-19/variants-concern.
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

