Proteomics Disclose the Potential of Gingival Crevicular Fluid (GCF) as a Source of Biomarkers for Severe Periodontitis
- PMID: 35329612
- PMCID: PMC8950923
- DOI: 10.3390/ma15062161
Proteomics Disclose the Potential of Gingival Crevicular Fluid (GCF) as a Source of Biomarkers for Severe Periodontitis
Abstract
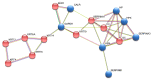
Periodontal disease is a widespread disorder comprising gingivitis, a mild early gum inflammation, and periodontitis, a more severe multifactorial inflammatory disease that, if left untreated, can lead to the gradual destruction of the tooth-supporting apparatus. To date, effective etiopathogenetic models fully explaining the clinical features of periodontal disease are not available. Obviously, a better understanding of periodontal disease could facilitate its diagnosis and improve its treatment. The purpose of this study was to employ a proteomic approach to analyze the gingival crevicular fluid (GCF) of patients with severe periodontitis, in search of potential biomarkers. GCF samples, collected from both periodontally healthy sites (H-GCF) and the periodontal pocket (D-GCF), were subjected to a comparison analysis using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). A total of 26 significantly different proteins, 14 up-regulated and 12 down-regulated in D-GCF vs. H-GCF, were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The main expressed proteins were inflammatory molecules, immune responders, and host enzymes. Most of these proteins were functionally connected using the STRING analysis database. Once validated in a large scale-study, these proteins could represent a cluster of promising biomarkers capable of making a valuable contribution for a better assessment of periodontitis.
Keywords: SDS-PAGE; biomarkers; gingival crevicular fluid; mass spectrometry; periodontal disease; periodontitis; proteomics.
Conflict of interest statement
The authors declare no conflict of interest related to this study.
Figures

Similar articles
-
A new albumin-depletion strategy improves proteomic research of gingival crevicular fluid from periodontitis patients.Clin Oral Investig. 2018 Apr;22(3):1375-1384. doi: 10.1007/s00784-017-2213-0. Epub 2017 Oct 9. Clin Oral Investig. 2018. PMID: 28993918
-
Liquid chromatography method with tandem mass spectrometry and fluorescence detection for determination of inflammatory biomarkers in gingival crevicular fluid as a tool for diagnosis of periodontal disease.J Pharm Biomed Anal. 2022 Apr 1;212:114644. doi: 10.1016/j.jpba.2022.114644. Epub 2022 Feb 4. J Pharm Biomed Anal. 2022. PMID: 35151070
-
Identification of azurocidin as a potential periodontitis biomarker by a proteomic analysis of gingival crevicular fluid.Proteome Sci. 2011 Jul 28;9:42. doi: 10.1186/1477-5956-9-42. Proteome Sci. 2011. PMID: 21794177 Free PMC article.
-
Current Status of Proteomic Technologies for Discovering and Identifying Gingival Crevicular Fluid Biomarkers for Periodontal Disease.Int J Mol Sci. 2018 Dec 26;20(1):86. doi: 10.3390/ijms20010086. Int J Mol Sci. 2018. PMID: 30587811 Free PMC article. Review.
-
The emerging role of small extracellular vesicles in saliva and gingival crevicular fluid as diagnostics for periodontitis.J Periodontal Res. 2022 Jan;57(1):219-231. doi: 10.1111/jre.12950. Epub 2021 Nov 13. J Periodontal Res. 2022. PMID: 34773636 Review.
Cited by
-
Characterisation of the periodontal proteome in gingival crevicular fluid and saliva using SWATH-MS.Front Cell Infect Microbiol. 2025 May 2;15:1576906. doi: 10.3389/fcimb.2025.1576906. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40384977 Free PMC article.
-
The Potential of Gingival Crevicular Fluid as a Tool for Molecular Diagnosis: A Systematic Review.Biomed Res Int. 2024 Oct 15;2024:5560866. doi: 10.1155/2024/5560866. eCollection 2024. Biomed Res Int. 2024. PMID: 39445210 Free PMC article.
-
Impact of smoking habit on the subgingival proteome in patients with periodontitis.J Periodontol. 2025 Mar;96(3):217-229. doi: 10.1002/JPER.24-0062. Epub 2024 Sep 16. J Periodontol. 2025. PMID: 39282712 Free PMC article.
-
Oral and Gingival Crevicular Fluid Biomarkers for Jawbone Turnover Diseases: A Scoping Review.Diagnostics (Basel). 2024 Sep 30;14(19):2184. doi: 10.3390/diagnostics14192184. Diagnostics (Basel). 2024. PMID: 39410587 Free PMC article.
-
The salivary metabolomics analyses reveal the variable metabolites in distinct staging of periodontitis.BMC Oral Health. 2025 Apr 3;25(1):480. doi: 10.1186/s12903-025-05792-y. BMC Oral Health. 2025. PMID: 40181365 Free PMC article.
References
-
- Papapanou P.N., Sanz M., Buduneli N., Dietrich T., Feres M., Fine D.H., Flemmig T.F., Garcia R., Giannobile W.V., Graziani F., et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018;89:S173–S182. doi: 10.1002/JPER.17-0721. - DOI - PubMed
-
- Khurshid Z., Khan E. Future of oral proteomics. J. Oral Res. 2018;7:42–43. doi: 10.17126/joralres.2018.019. - DOI
-
- Nisha K.J., Annie K.G. Proteomics–The future of periodontal diagnostics. Biomed. J. Sci. Tech. Res. 2017;1:1402–1406. doi: 10.26717/BJSTR.2017.01.000447. - DOI
LinkOut - more resources
Full Text Sources

