Multigene Phylogeny Reveals Endophytic Xylariales Novelties from Dendrobium Species from Southwestern China and Northern Thailand
- PMID: 35330250
- PMCID: PMC8955275
- DOI: 10.3390/jof8030248
Multigene Phylogeny Reveals Endophytic Xylariales Novelties from Dendrobium Species from Southwestern China and Northern Thailand
Abstract
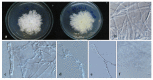
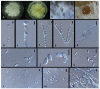
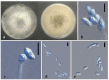
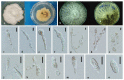
Xylariales are common endophytes of Dendrobium. However, xylarialean species resolution remains difficult without sequence data and poor sporulation on artificial media and asexual descriptions for only several species and old type material. The surface-sterilized and morph-molecular methods were used for fungal isolation and identification. A total of forty-seven strains were identified as twenty-three species belonging to Apiosporaceae, Hypoxylaceae, Induratiaceae, and Xylariaceae. Five new species-Annulohypoxylon moniliformis, Apiospora dendrobii, Hypoxylon endophyticum, H. officinalis and Nemania dendrobii were discovered. Three tentative new species were speculated in Xylaria. Thirteen known fungal species from Hypoxylon, Nemania, Nigrospora, and Xylaria were also identified. Another two strains were only identified at the genus and family level (Induratia sp., Hypoxylaceae sp.). This study recorded 12 new hosts for xylarialean endophytes. This is the first report of Xylariales species as endophytes from Dendrobium aurantiacum var. denneanum, D. cariniferum, D. harveyanum, D. hercoglossum, D. moniliforme, and D. moschatum. Dendrobium is associated with abundant xylarialean taxa, especially species of Hypoxylon and Xylaria. We recommend the use of oat agar with low concentrations to induce sporulation of Xylaria strains.
Keywords: Xylariomycetidae; endophytes; multi-locus phylogeny; oat media; orchids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




















Similar articles
-
Endophytic Colletotrichum species from Dendrobium spp. in China and Northern Thailand.MycoKeys. 2018 Dec 4;(43):23-57. doi: 10.3897/mycokeys.43.25081. eCollection 2018. MycoKeys. 2018. PMID: 30568535 Free PMC article.
-
Redisposition of apiosporous genera Induratia and Muscodor in the Xylariales, following the discovery of an authentic strain of Induratia apiospora.Bot Stud. 2023 Apr 13;64(1):8. doi: 10.1186/s40529-023-00372-1. Bot Stud. 2023. PMID: 37052736 Free PMC article.
-
Novel discoveries of Xylariomycetidae (Ascomycota) taxa from peat swamp forests and other terrestrial habitats in Thailand.MycoKeys. 2024 Aug 7;107:219-247. doi: 10.3897/mycokeys.107.127749. eCollection 2024. MycoKeys. 2024. PMID: 39169992 Free PMC article.
-
Secondary metabolites of endophytic Xylaria species with potential applications in medicine and agriculture.World J Microbiol Biotechnol. 2017 Jan;33(1):15. doi: 10.1007/s11274-016-2174-5. Epub 2016 Nov 28. World J Microbiol Biotechnol. 2017. PMID: 27896581 Review.
-
Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales.Nat Prod Rep. 2018 Sep 19;35(9):992-1014. doi: 10.1039/c8np00010g. Nat Prod Rep. 2018. PMID: 29774351 Review.
Cited by
-
Endophytic xylariaceous fungi from rice in northern Thailand: discovery of novel species and new host records.MycoKeys. 2025 Jul 29;120:1-34. doi: 10.3897/mycokeys.120.152187. eCollection 2025. MycoKeys. 2025. PMID: 40778232 Free PMC article.
-
Study on endolichenic fungal assemblage in Parmotrema and Heterodermia lichens of Shivamoga, Karnataka.Mol Biol Rep. 2024 Apr 20;51(1):549. doi: 10.1007/s11033-024-09497-3. Mol Biol Rep. 2024. PMID: 38642168
-
Fungal communities associated with early immature tubers of wild Gastrodia elata.Ecol Evol. 2024 Feb 21;14(2):e11004. doi: 10.1002/ece3.11004. eCollection 2024 Feb. Ecol Evol. 2024. PMID: 38389997 Free PMC article.
-
Morphology and multigene phylogeny reveal four new Xylaria (Xylariales, Xylariaceae) species from karst region in China.MycoKeys. 2024 Sep 4;108:169-196. doi: 10.3897/mycokeys.108.130565. eCollection 2024. MycoKeys. 2024. PMID: 39268505 Free PMC article.
-
Four New Endophytic Apiospora Species Isolated from Three Dicranopteris Species in Guizhou, China.J Fungi (Basel). 2023 Nov 10;9(11):1096. doi: 10.3390/jof9111096. J Fungi (Basel). 2023. PMID: 37998901 Free PMC article.
References
-
- Moudi M., Go R. Morphological study of four sections of genus Dendrobium SW.(Orchidaceae) in Peninsular Malaysia. Pak. J. Bot. 2017;49:569–577.
-
- Wu M., Shu Y., Song L., Liu B., Zhang L., Wang L., Liu Y., Bi J., Xiong C., Cao Z., et al. Prenatal exposure to thallium is associated with decreased mitochondrial DNA copy number in newborns: Evidence from a birth cohort study. Environ. Int. 2019;129:470–477. doi: 10.1016/j.envint.2019.05.053. - DOI - PubMed
-
- Kabir M., Rahman M., Jamal A., Rahman M., Khalekuzzaman M. Multiple shoot regeneration in Dendrobium fimbriatum Hook an ornamental orchid. J. Anim. Plant Sci. 2013;23:1140–1145.
-
- Fracchia S., Aranda-Rickert A., Rothen C., Sede S. Associated fungi, symbiotic germination and in vitro seedling development of the rare Andean terrestrial orchid Chloraea riojana. Flora. 2016;224:106–111. doi: 10.1016/j.flora.2016.07.008. - DOI
LinkOut - more resources
Full Text Sources

