Phylogenomics and Comparative Genomics Highlight Specific Genetic Features in Ganoderma Species
- PMID: 35330313
- PMCID: PMC8955403
- DOI: 10.3390/jof8030311
Phylogenomics and Comparative Genomics Highlight Specific Genetic Features in Ganoderma Species
Abstract
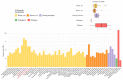
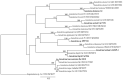
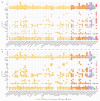
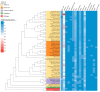
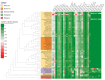
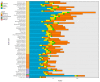
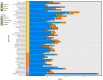
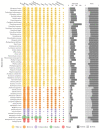
The Ganoderma species in Polyporales are ecologically and economically relevant wood decayers used in traditional medicine, but their genomic traits are still poorly documented. In the present study, we carried out a phylogenomic and comparative genomic analyses to better understand the genetic blueprint of this fungal lineage. We investigated seven Ganoderma genomes, including three new genomes, G. australe, G. leucocontextum, and G. lingzhi. The size of the newly sequenced genomes ranged from 60.34 to 84.27 Mb and they encoded 15,007 to 20,460 genes. A total of 58 species, including 40 white-rot fungi, 11 brown-rot fungi, four ectomycorrhizal fungi, one endophyte fungus, and two pathogens in Basidiomycota, were used for phylogenomic analyses based on 143 single-copy genes. It confirmed that Ganoderma species belong to the core polyporoid clade. Comparing to the other selected species, the genomes of the Ganoderma species encoded a larger set of genes involved in terpene metabolism and coding for secreted proteins (CAZymes, lipases, proteases and SSPs). Of note, G. australe has the largest genome size with no obvious genome wide duplication, but showed transposable elements (TEs) expansion and the largest set of terpene gene clusters, suggesting a high ability to produce terpenoids for medicinal treatment. G. australe also encoded the largest set of proteins containing domains for cytochrome P450s, heterokaryon incompatibility and major facilitator families. Besides, the size of G. australe secretome is the largest, including CAZymes (AA9, GH18, A01A), proteases G01, and lipases GGGX, which may enhance the catabolism of cell wall carbohydrates, proteins, and fats during hosts colonization. The current genomic resource will be used to develop further biotechnology and medicinal applications, together with ecological studies of the Ganoderma species.
Keywords: Ganoderma; genomics; secondary metabolism; secretome; terpenes; wood decay.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Rodríguez-Couto S. Industrial an environmental application of white-rot fungi. Mycosphere. 2017;8:456–466. doi: 10.5943/mycosphere/8/3/7. - DOI
-
- Seman I.B. Ph.D. Thesis. Universiti Putra Malaysia; Selangor, Malaysia: 2018. R&D on Biology, Detection and Management of Ganoderma Disease in Oil Palm.
-
- Wang H., Deng W., Shen M., Yan G., Zhao W., Yang Y. A laccase Gl-LAC-4 purified from white-rot fungus Ganoderma lucidum had a strong ability to degrade and detoxify the alkylphenol pollutants 4-n-octylphenol and 2-phenylphenol. J. Hazard. Mater. 2021;408:124775. doi: 10.1016/j.jhazmat.2020.124775. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

