MEDUSA: A Pipeline for Sensitive Taxonomic Classification and Flexible Functional Annotation of Metagenomic Shotgun Sequences
- PMID: 35330728
- PMCID: PMC8940201
- DOI: 10.3389/fgene.2022.814437
MEDUSA: A Pipeline for Sensitive Taxonomic Classification and Flexible Functional Annotation of Metagenomic Shotgun Sequences
Abstract
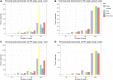
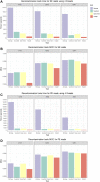
Metagenomic studies unravel details about the taxonomic composition and the functions performed by microbial communities. As a complete metagenomic analysis requires different tools for different purposes, the selection and setup of these tools remain challenging. Furthermore, the chosen toolset will affect the accuracy, the formatting, and the functional identifiers reported in the results, impacting the results interpretation and the biological answer obtained. Thus, we surveyed state-of-the-art tools available in the literature, created simulated datasets, and performed benchmarks to design a sensitive and flexible metagenomic analysis pipeline. Here we present MEDUSA, an efficient pipeline to conduct comprehensive metagenomic analyses. It performs preprocessing, assembly, alignment, taxonomic classification, and functional annotation on shotgun data, supporting user-built dictionaries to transfer annotations to any functional identifier. MEDUSA includes several tools, as fastp, Bowtie2, DIAMOND, Kaiju, MEGAHIT, and a novel tool implemented in Python to transfer annotations to BLAST/DIAMOND alignment results. These tools are installed via Conda, and the workflow is managed by Snakemake, easing the setup and execution. Compared with MEGAN 6 Community Edition, MEDUSA correctly identifies more species, especially the less abundant, and is more suited for functional analysis using Gene Ontology identifiers.
Keywords: bioinformatics; functional annotation; metagenomics; pipeline; shotgun sequences; taxonomic classification.
Copyright © 2022 Morais, Cavalcante, Monteiro, Pasquali and Dalmolin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
MetaLAFFA: a flexible, end-to-end, distributed computing-compatible metagenomic functional annotation pipeline.BMC Bioinformatics. 2020 Oct 21;21(1):471. doi: 10.1186/s12859-020-03815-9. BMC Bioinformatics. 2020. PMID: 33087062 Free PMC article.
-
Sunbeam: an extensible pipeline for analyzing metagenomic sequencing experiments.Microbiome. 2019 Mar 22;7(1):46. doi: 10.1186/s40168-019-0658-x. Microbiome. 2019. PMID: 30902113 Free PMC article.
-
Introduction to the Analysis of Environmental Sequences: Metagenomics with MEGAN.Methods Mol Biol. 2019;1910:591-604. doi: 10.1007/978-1-4939-9074-0_19. Methods Mol Biol. 2019. PMID: 31278678
-
HOME-BIO (sHOtgun MEtagenomic analysis of BIOlogical entities): a specific and comprehensive pipeline for metagenomic shotgun sequencing data analysis.BMC Bioinformatics. 2021 Jul 5;22(Suppl 7):106. doi: 10.1186/s12859-021-04004-y. BMC Bioinformatics. 2021. PMID: 34225648 Free PMC article. Review.
-
Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences.Brief Bioinform. 2020 May 21;21(3):777-790. doi: 10.1093/bib/bbz025. Brief Bioinform. 2020. PMID: 30860572 Free PMC article. Review.
Cited by
-
Mock community taxonomic classification performance of publicly available shotgun metagenomics pipelines.Sci Data. 2024 Jan 17;11(1):81. doi: 10.1038/s41597-023-02877-7. Sci Data. 2024. PMID: 38233447 Free PMC article.
-
Comprehensive genomics, probiotic, and antibiofilm potential analysis of Streptococcus thermophilus strains isolated from homemade and commercial dahi.Sci Rep. 2025 Feb 27;15(1):7089. doi: 10.1038/s41598-025-90999-w. Sci Rep. 2025. PMID: 40016393 Free PMC article.
-
Metagenomic Analyses Reveal the Influence of Depth Layers on Marine Biodiversity on Tropical and Subtropical Regions.Microorganisms. 2023 Jun 27;11(7):1668. doi: 10.3390/microorganisms11071668. Microorganisms. 2023. PMID: 37512841 Free PMC article.
References
-
- Babraham (2021). FastQC. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed Oct 07, 2021).
-
- BBTools (2021). BBTools. Available at: http://jgi.doe.gov/data-and-tools/bb-tools/ (Accessed Oct 07, 2021).
LinkOut - more resources
Full Text Sources
Research Materials

