Dissecting the Species-Specific Virome in Culicoides of Thrace
- PMID: 35330767
- PMCID: PMC8940260
- DOI: 10.3389/fmicb.2022.802577
Dissecting the Species-Specific Virome in Culicoides of Thrace
Abstract
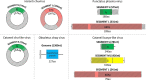
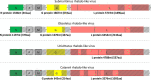
Biting midges (Culicoides) are vectors of arboviruses of both veterinary and medical importance. The surge of emerging and reemerging vector-borne diseases and their expansion in geographical areas affected by climate change has increased the importance of understanding their capacity to contribute to novel and emerging infectious diseases. The study of Culicoides virome is the first step in the assessment of this potential. In this study, we analyzed the RNA virome of 10 Culicoides species within the geographical area of Thrace in the southeastern part of Europe, a crossing point between Asia and Europe and important path of various arboviruses, utilizing the Ion Torrent next-generation sequencing (NGS) platform and a custom bioinformatics pipeline based on TRINITY assembler and alignment algorithms. The analysis of the RNA virome of 10 Culicoides species resulted in the identification of the genomic signatures of 14 novel RNA viruses, including three fully assembled viruses and four segmented viruses with at least one segment fully assembled, most of which were significantly divergent from previously identified related viruses from the Solemoviridae, Phasmaviridae, Phenuiviridae, Reoviridae, Chuviridae, Partitiviridae, Orthomyxoviridae, Rhabdoviridae, and Flaviviridae families. Each Culicoides species carried a species-specific set of viruses, some of which are related to viruses from other insect vectors in the same area, contributing to the idea of a virus-carrier web within the ecosystem. The identified viruses not only expand our current knowledge on the virome of Culicoides but also set the basis of the genetic diversity of such viruses in the area of southeastern Europe. Furthermore, our study highlights that such metagenomic approaches should include as many species as possible of the local virus-carrier web that interact and share the virome of a geographical area.
Keywords: Culicoides; Greece; Thrace; arboviruses; emerging infectious diseases; metagenomics; vectors; virome analysis.
Copyright © 2022 Konstantinidis, Bampali, de Courcy Williams, Dovrolis, Gatzidou, Papazilakis, Nearchou, Veletza and Karakasiliotis.
Conflict of interest statement
PP was employed by the company Evrofarma S.A. AN was employed by the company Geotechno Ygeionomiki O.E. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources

