Context dependent isoform specific PI3K inhibition confers drug resistance in hepatocellular carcinoma cells
- PMID: 35331184
- PMCID: PMC8953069
- DOI: 10.1186/s12885-022-09357-y
Context dependent isoform specific PI3K inhibition confers drug resistance in hepatocellular carcinoma cells
Abstract
Background: Targeted therapies for Primary liver cancer (HCC) is limited to the multi-kinase inhibitors, and not fully effective due to the resistance to these agents because of the heterogeneous molecular nature of HCC developed during chronic liver disease stages and cirrhosis. Although combinatorial therapy can increase the efficiency of targeted therapies through synergistic activities, isoform specific effects of the inhibitors are usually ignored. This study concentrated on PI3K/Akt/mTOR pathway and the differential combinatory bioactivities of isoform specific PI3K-α inhibitor (PIK-75) or PI3K-β inhibitor (TGX-221) with Sorafenib dependent on PTEN context.
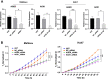
Methods: The bioactivities of inhibitors on PTEN adequate Huh7 and deficient Mahlavu cells were investigated with real time cell growth, cell cycle and cell migration assays. Differentially expressed genes from RNA-Seq were identified by edgeR tool. Systems level network analysis of treatment specific pathways were performed with Prize Collecting Steiner Tree (PCST) on human interactome and enriched networks were visualized with Cytoscape platform.
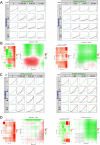
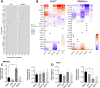
Results: Our data from combinatory treatment of Sorafenib and PIK-75 and TGX-221 showed opposite effects; while PIK-75 displays synergistic effects on Huh7 cells leading to apoptotic cell death, Sorafenib with TGX-221 display antagonistic effects and significantly promotes cell growth in PTEN deficient Mahlavu cells. Signaling pathways were reconstructed and analyzed in-depth from RNA-Seq data to understand mechanism of differential synergistic or antagonistic effects of PI3K-α (PIK-75) and PI3K-β (TGX-221) inhibitors with Sorafenib. PCST allowed as to identify AOX1 and AGER as targets in PI3K/Akt/mTOR pathway for this combinatory effect. The siRNA knockdown of AOX1 and AGER significantly reduced cell proliferation in HCC cells.
Conclusions: Simultaneously constructed and analyzed differentially expressed cellular networks presented in this study, revealed distinct consequences of isoform specific PI3K inhibition in PTEN adequate and deficient liver cancer cells. We demonstrated the importance of context dependent and isoform specific PI3K/Akt/mTOR signaling inhibition in drug resistance during combination therapies. ( https://github.com/cansyl/Isoform-spesific-PI3K-inhibitor-analysis ).
Keywords: Liver Cancer; Network analysis; PI3K/Akt/mTOR pathway; Resistance; Synergy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

