A copper switch for inducing CRISPR/Cas9-based transcriptional activation tightly regulates gene expression in Nicotiana benthamiana
- PMID: 35331211
- PMCID: PMC8943966
- DOI: 10.1186/s12896-022-00741-x
A copper switch for inducing CRISPR/Cas9-based transcriptional activation tightly regulates gene expression in Nicotiana benthamiana
Abstract
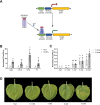
Background: CRISPR-based programmable transcriptional activators (PTAs) are used in plants for rewiring gene networks. Better tuning of their activity in a time and dose-dependent manner should allow precise control of gene expression. Here, we report the optimization of a Copper Inducible system called CI-switch for conditional gene activation in Nicotiana benthamiana. In the presence of copper, the copper-responsive factor CUP2 undergoes a conformational change and binds a DNA motif named copper-binding site (CBS).
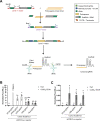
Results: In this study, we tested several activation domains fused to CUP2 and found that the non-viral Gal4 domain results in strong activation of a reporter gene equipped with a minimal promoter, offering advantages over previous designs. To connect copper regulation with downstream programmable elements, several copper-dependent configurations of the strong dCasEV2.1 PTA were assayed, aiming at maximizing activation range, while minimizing undesired background expression. The best configuration involved a dual copper regulation of the two protein components of the PTA, namely dCas9:EDLL and MS2:VPR, and a constitutive RNA pol III-driven expression of the third component, a guide RNA with anchoring sites for the MS2 RNA-binding domain. With these optimizations, the CI/dCasEV2.1 system resulted in copper-dependent activation rates of 2,600-fold and 245-fold for the endogenous N. benthamiana DFR and PAL2 genes, respectively, with negligible expression in the absence of the trigger.
Conclusions: The tight regulation of copper over CI/dCasEV2.1 makes this system ideal for the conditional production of plant-derived metabolites and recombinant proteins in the field.
Keywords: CRISPR/Cas9-based programmable transcriptional activator; Copper switch; Inducible expression; Minimal promoter; Nicotiana benthamiana; Plant synthetic biology.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Boetti H, Chevalier L, Denmat LA, Thomas D, Thomasset B. Efficiency of physical (light) or chemical (ABA, tetracycline, CuSO4 or 2-CBSU)-stimulus-dependent gus gene expression in tobacco cell suspensions. Biotechnol Bioeng. 1999;64(1):1–13. - PubMed
-
- Burkhead JL, Gogolin Reynolds KA, Abdel-Ghany SE, Cohu CM, Pilon M. Copper homeostasis. New Phytol. 2009;182(4):799–816. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

