Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels
- PMID: 35331218
- PMCID: PMC8944098
- DOI: 10.1186/s12915-022-01277-4
Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels
Abstract
Background: Extracellular vesicles (EVs) are released by nearly every cell type and have attracted much attention for their ability to transfer protein and diverse RNA species from donor to recipient cells. Much attention has been given so far to the features of EV short RNAs such as miRNAs. However, while the presence of mRNA and long noncoding RNA (lncRNA) transcripts in EVs has also been reported by multiple different groups, the properties and function of these longer transcripts have been less thoroughly explored than EV miRNA. Additionally, the impact of EV export on the transcriptome of exporting cells has remained almost completely unexamined. Here, we globally investigate mRNA and lncRNA transcripts in endothelial EVs in multiple different conditions.
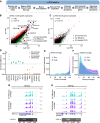
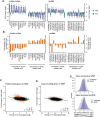
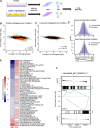
Results: In basal conditions, long RNA transcripts enriched in EVs have longer than average half-lives and distinctive stability-related sequence and structure characteristics including shorter transcript length, higher exon density, and fewer 3' UTR A/U-rich elements. EV-enriched long RNA transcripts are also enriched in HNRNPA2B1 binding motifs and are impacted by HNRNPA2B1 depletion, implicating this RNA-binding protein in the sorting of long RNA to EVs. After signaling-dependent modification of the cellular transcriptome, we observed that, unexpectedly, the rate of EV enrichment relative to cells was altered for many mRNA and lncRNA transcripts. This change in EV enrichment was negatively correlated with intracellular abundance, with transcripts whose export to EVs increased showing decreased abundance in cells and vice versa. Correspondingly, after treatment with inhibitors of EV secretion, levels of mRNA and lncRNA transcripts that are normally highly exported to EVs increased in cells, indicating a measurable impact of EV export on the long RNA transcriptome of the exporting cells. Compounds with different mechanisms of inhibition of EV secretion affected the cellular transcriptome differently, suggesting the existence of multiple EV subtypes with different long RNA profiles.
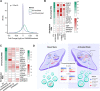
Conclusions: We present evidence for an impact of EV physiology on the characteristics of EV-producing cell transcriptomes. Our work suggests a new paradigm in which the sorting and packaging of transcripts into EVs participate, together with transcription and RNA decay, in controlling RNA homeostasis and shape the cellular long RNA abundance profile.
Keywords: Exosomes; Extracellular vesicles; Gene regulation; HNRNPA2B1; lncRNA; mRNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

