The pathogens of secondary infection in septic patients share a similar genotype to those that predominate in the gut
- PMID: 35331299
- PMCID: PMC8944137
- DOI: 10.1186/s13054-022-03943-z
The pathogens of secondary infection in septic patients share a similar genotype to those that predominate in the gut
Abstract
Background: Secondary nosocomial infections, which are commonly caused by carbapenem-resistant Klebsiella pneumoniae (CRKP) and vancomycin-resistant Enterococcus faecium (VRE), often develop in septic patients. This study aimed to identify the origin of secondary systemic pathogens and reveal the underlying mechanism of infection.
Methods: In this prospective, observational case-control study, a total of 34 septic patients, 33 non-septic intensive care unit (ICU) patients and 10 healthy individuals serving as controls were enrolled. Three hundred and twelve fecal samples were collected and subjected to 16S rRNA gene amplicon sequencing. Metagenome sequencing was performed to identify the homology between dominant CRKP or VRE in the intestine and pathogens isolated from secondary infectious sites. C57/BL mice were established as pseudo germ-free animal model by pretreatment with broad-spectrum antibiotics for two weeks.
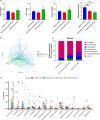
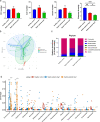
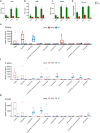
Results: The abundance and diversity of the gut microbiota in septic patients was drastically decreased one week after ICU admission, potentially leading to the enrichment of antibiotic-resistant bacteria, such as CRKP. Furthermore, secondary bloodstream and abdominal infections caused by CRKP or VRE in septic patients occurred after intestinal colonization with the predominant bacterial species. Genomic analysis showed that bacteria isolated from secondary infection had high homology with the corresponding predominant intestinal opportunistic pathogens. In addition, animal model experiments validated the hypothesis that the administration of antibiotics caused the enrichment of CRKP and VRE among the intestinal microbiota, increasing the likelihood of permeation of other tissues and potentially causing subsequent systemic infection in pseudo germ-free mice.
Conclusion: Our study indicated that the pathogens causing secondary infection in septic patients might originate from the intestinal colonization of pathogens following broad-spectrum antibiotic treatment.
Keywords: Bacterial translocation; Gut microbiota; Secondary nosocomial infection; Sepsis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Deutschman CS, Tracey KJ. Sepsis: current dogma and new perspectives. Immunity. 2014;40(4):463–475. - PubMed
-
- Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348(16):1546–1554. - PubMed
-
- Haak BW, Wiersinga WJ. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol. 2017;2(2):135–143. - PubMed
-
- Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

