Research Techniques Made Simple: Spatial Transcriptomics
- PMID: 35331388
- PMCID: PMC8969263
- DOI: 10.1016/j.jid.2021.12.014
Research Techniques Made Simple: Spatial Transcriptomics
Abstract
Transcriptome profiling of tissues and single cells facilitates interrogation of gene expression changes within diverse biological contexts. However, spatial information is often lost during tissue homogenization or dissociation. Recent advances in transcriptome profiling preserve the in situ spatial contexts of RNA molecules and together comprise a group of techniques known as spatial transcriptomics (ST), enabling localization of cell types and their associated gene expression within intact tissues. In this paper, we review ST methods; summarize data analysis approaches, including integration with single-cell transcriptomics data; and discuss their applications in dermatologic research. These tools offer a promising avenue toward improving our understanding of niche patterning and cell‒cell interactions within heterogeneous tissues that encompass skin homeostasis and disease.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest
The authors declare no conflicts of interest.
Figures
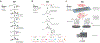


References
-
- Asp M, Bergenstråhle J, Lundeberg J. Spatially Resolved Transcriptomes—Next Generation Tools for Tissue Exploration. BioEssays. 2020;42(10):1–16 - PubMed
-
- Bergenstråhle L, He B, Bergenstråhle J, Andersson A, Lundeberg J, Zou J, et al. Super-resolved spatial transcriptomics by deep data fusion. bioRxiv. 2020;2020.02.28.963413 Available from: https://www.biorxiv.org/content/10.1101/2020.02.28.963413v1.full.pdf+html - DOI - PubMed
-
- Van Den Brink SC, Sage F, Vértesy Á, Spanjaard B, Peterson-Maduro J, Baron CS, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 2017;14(10):935–6 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

