Notch signaling pathway: architecture, disease, and therapeutics
- PMID: 35332121
- PMCID: PMC8948217
- DOI: 10.1038/s41392-022-00934-y
Notch signaling pathway: architecture, disease, and therapeutics
Abstract
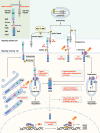
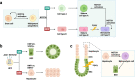
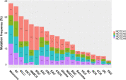
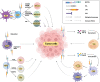
The NOTCH gene was identified approximately 110 years ago. Classical studies have revealed that NOTCH signaling is an evolutionarily conserved pathway. NOTCH receptors undergo three cleavages and translocate into the nucleus to regulate the transcription of target genes. NOTCH signaling deeply participates in the development and homeostasis of multiple tissues and organs, the aberration of which results in cancerous and noncancerous diseases. However, recent studies indicate that the outcomes of NOTCH signaling are changeable and highly dependent on context. In terms of cancers, NOTCH signaling can both promote and inhibit tumor development in various types of cancer. The overall performance of NOTCH-targeted therapies in clinical trials has failed to meet expectations. Additionally, NOTCH mutation has been proposed as a predictive biomarker for immune checkpoint blockade therapy in many cancers. Collectively, the NOTCH pathway needs to be integrally assessed with new perspectives to inspire discoveries and applications. In this review, we focus on both classical and the latest findings related to NOTCH signaling to illustrate the history, architecture, regulatory mechanisms, contributions to physiological development, related diseases, and therapeutic applications of the NOTCH pathway. The contributions of NOTCH signaling to the tumor immune microenvironment and cancer immunotherapy are also highlighted. We hope this review will help not only beginners but also experts to systematically and thoroughly understand the NOTCH signaling pathway.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Figures were created with biorender.com.
Figures

References
-
- Yochem J, Weston K, Greenwald I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. 1988;335:547–550. - PubMed
-
- Austin J, Kimble J. Transcript analysis of glp-1 and lin-12, homologous genes required for cell interactions during development of C. elegans. Cell. 1989;58:565–571. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

