Activation of Drp1 promotes fatty acids-induced metabolic reprograming to potentiate Wnt signaling in colon cancer
- PMID: 35332310
- PMCID: PMC9525627
- DOI: 10.1038/s41418-022-00974-5
Activation of Drp1 promotes fatty acids-induced metabolic reprograming to potentiate Wnt signaling in colon cancer
Abstract
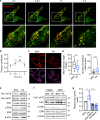
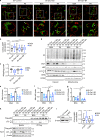
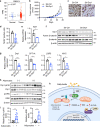
Cancer cells are known for their ability to adapt variable metabolic programs depending on the availability of specific nutrients. Our previous studies have shown that uptake of fatty acids alters cellular metabolic pathways in colon cancer cells to favor fatty acid oxidation. Here, we show that fatty acids activate Drp1 to promote metabolic plasticity in cancer cells. Uptake of fatty acids (FAs) induces mitochondrial fragmentation by promoting ERK-dependent phosphorylation of Drp1 at the S616 site. This increased phosphorylation of Drp1 enhances its dimerization and interaction with Mitochondrial Fission Factor (MFF) at the mitochondria. Consequently, knockdown of Drp1 or MFF attenuates fatty acid-induced mitochondrial fission. In addition, uptake of fatty acids triggers mitophagy via a Drp1- and p62-dependent mechanism to protect mitochondrial integrity. Moreover, results from metabolic profiling analysis reveal that silencing Drp1 disrupts cellular metabolism and blocks fatty acid-induced metabolic reprograming by inhibiting fatty acid utilization. Functionally, knockdown of Drp1 decreases Wnt/β-catenin signaling by preventing fatty acid oxidation-dependent acetylation of β-catenin. As a result, Drp1 depletion inhibits the formation of tumor organoids in vitro and xenograft tumor growth in vivo. Taken together, our study identifies Drp1 as a key mediator that connects mitochondrial dynamics with fatty acid metabolism and cancer cell signaling.
© 2022. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

