Bound nucleotide can control the dynamic architecture of monomeric actin
- PMID: 35332323
- PMCID: PMC9010300
- DOI: 10.1038/s41594-022-00743-5
Bound nucleotide can control the dynamic architecture of monomeric actin
Abstract
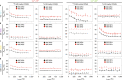
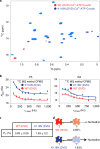
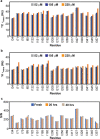
Polymerization of actin into cytoskeletal filaments is coupled to its bound adenine nucleotides. The mechanism by which nucleotide modulates actin functions has not been evident from analyses of ATP- and ADP-bound crystal structures of the actin monomer. We report that NMR chemical shift differences between the two forms are globally distributed. Furthermore, microsecond-millisecond motions are spread throughout the molecule in the ATP form, but largely confined to subdomains 1 and 2, and the nucleotide binding site in the ADP form. Through these motions, the ATP- and ADP-bound forms sample different high-energy conformations. A deafness-causing, fast-nucleating actin mutant populates the high-energy conformer of ATP-actin more than the wild-type protein, suggesting that this conformer may be on the pathway to nucleation. Together, the data suggest a model in which differential sampling of a nucleation-compatible form of the actin monomer may contribute to control of actin filament dynamics by nucleotide.
© 2022. The Author(s).
Conflict of interest statement
M.K.R. is a founder of Faze Medicines. R.A. and J.A.Z. declare no competing interests.
Figures










References
-
- Oosawa, F. & Asakura, S. Thermodynamics of the Polymerization of Protein (Academic, 1975).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

