Re-evaluation of missense variant classifications in NF2
- PMID: 35332608
- PMCID: PMC9323416
- DOI: 10.1002/humu.24370
Re-evaluation of missense variant classifications in NF2
Abstract
Missense variants in the NF2 gene result in variable NF2 disease presentation. Clinical classification of missense variants often represents a challenge, due to lack of evidence for pathogenicity and function. This study provides a summary of NF2 missense variants, with variant classifications based on currently available evidence. NF2 missense variants were collated from pathology-associated databases and existing literature. Association for Clinical Genomic Sciences Best Practice Guidelines (2020) were followed in the application of evidence for variant interpretation and classification. The majority of NF2 missense variants remain classified as variants of uncertain significance. However, NF2 missense variants identified in gnomAD occurred at a consistent rate across the gene, while variants compiled from pathology-associated databases displayed differing rates of variation by exon of NF2. The highest rate of NF2 disease-associated variants was observed in exon 7, while lower rates were observed toward the C-terminus of the NF2 protein, merlin. Further phenotypic information associated with variants, alongside variant-specific functional analysis, is necessary for more definitive variant interpretation. Our data identified differences in frequency of NF2 missense variants by exon between gnomAD population data and NF2 disease-associated variants, suggesting a potential genotype-phenotype correlation; further work is necessary to substantiate this.
Keywords: NF2; classification guidelines; missense; neurofibromatosis type 2; variant classification.
© 2022 The Authors. Human Mutation published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
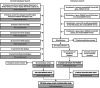
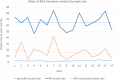

References
-
- Baser, M. E. , Kuramoto, L. , Woods, R. , Joe, H. , Friedman, J. M. , Wallace, A. J. , Ramsden, R. T. , Olschwang, S. , Bijlsma, E. , Kalamarides, M. , Papi, L. , Kato, R. , Carroll, J. , Lázaro, C. , Joncourt, F. , Parry, D. M. , Rouleau, G. A. , & Evans, D. G. (2005). The location of constitutional neurofibromatosis 2 (NF2) splice site mutations is associated with the severity of NF2. Journal of Medical Genetics, 42(7), 540–546. 10.1136/jmg.2004.029504 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

