Exosome Processing and Characterization Approaches for Research and Technology Development
- PMID: 35332686
- PMCID: PMC9130923
- DOI: 10.1002/advs.202103222
Exosome Processing and Characterization Approaches for Research and Technology Development
Abstract
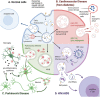
Exosomes are extracellular vesicles that share components of their parent cells and are attractive in biotechnology and biomedical research as potential disease biomarkers as well as therapeutic agents. Crucial to realizing this potential is the ability to manufacture high-quality exosomes; however, unlike biologics such as proteins, exosomes lack standardized Good Manufacturing Practices for their processing and characterization. Furthermore, there is a lack of well-characterized reference exosome materials to aid in selection of methods for exosome isolation, purification, and analysis. This review informs exosome research and technology development by comparing exosome processing and characterization methods and recommending exosome workflows. This review also provides a detailed introduction to exosomes, including their physical and chemical properties, roles in normal biological processes and in disease progression, and summarizes some of the on-going clinical trials.
Keywords: Good Manufacturing Practices; MISEV2018 guidelines; analytical characterizations; exosome clinical trials; exosomes; extracellular vesicles; isolation processes.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- a) Cheng H. L., Fu C. Y., Kuo W. C., Chen Y. W., Chen Y. S., Lee Y. M., Li K. H., Chen C. C., Ma H. P., Huang P. C., Wang Y. L., Lee G. B., Lab Chip 2018, 18, 2917; - PubMed
- b) Kura B., Kalocayova B., Devaux Y., Bartekova M., Int. J. Mol. Sci. 2020, 21, 700; - PMC - PubMed
- c) Osada‐Oka M., Shiota M., Izumi Y., Nishiyama M., Tanaka M., Yamaguchi T., Sakurai E., Miura K., Iwao H., Hypertens. Res. 2017, 40, 353. - PubMed
-
- a) Welch J. L., Stapleton J. T., Okeoma C. M., J. Gen. Virol. 2019, 100, 350; - PMC - PubMed
- b) Sims B., Farrow A. L., Williams S. D., Bansal A., Krendelchtchikov A., Matthews Q. L., Arch. Virol. 2018, 163, 1683; - PMC - PubMed
- c) Ouattara L. A., Anderson S. M., Doncel G. F., Andrologia 2018, 50, e13220; - PMC - PubMed
- d) Cheruiyot C., Pataki Z., Ramratnam B., Li M., Proteomics: Clin. Appl. 2018, 12, 1700142; - PubMed
- e) Pulliam L., Sun B., Mustapic M., Chawla S., Kapogiannis D., J. Neurovirol. 2019, 25, 702. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
