The Mnn10/Anp1-dependent N-linked outer chain glycan is dispensable for Candida albicans cell wall integrity
- PMID: 35333306
- PMCID: PMC9071539
- DOI: 10.1093/genetics/iyac048
The Mnn10/Anp1-dependent N-linked outer chain glycan is dispensable for Candida albicans cell wall integrity
Abstract
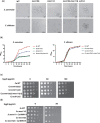
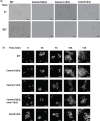
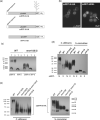
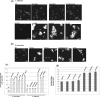
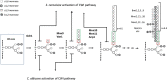
Candida albicans cell wall glycoproteins, and in particular their mannose-rich glycans, are important for maintaining cellular integrity as well as host recognition, adhesion, and immunomodulation. The asparagine (N)-linked mannose outer chain of these glycoproteins is produced by Golgi mannosyltransferases (MTases). The outer chain is composed of a linear backbone of ∼50 α1,6-linked mannoses, which acts as a scaffold for addition of ∼150 or more mannoses in other linkages. Here, we describe the characterization of C. albicans OCH1, MNN9, VAN1, ANP1, MNN10, and MNN11, which encode the conserved Golgi MTases that sequentially catalyze the α1,6 mannose outer chain backbone. Candida albicans och1Δ/Δ, mnn9Δ/Δ, and van1Δ/Δ mutants block the earliest steps of backbone synthesis and like their Saccharomyces cerevisiae counterparts, have severe cell wall and growth phenotypes. Unexpectedly, and in stark contrast to S. cerevisiae, loss of Anp1, Mnn10, or Mnn11, which together synthesize most of the backbone, have no obvious deleterious phenotypes. These mutants were unaffected in cell morphology, growth, drug sensitivities, hyphal formation, and macrophage recognition. Analyses of secreted glycosylation reporters demonstrated that anp1Δ/Δ, mnn10Δ/Δ, and mnn11Δ/Δ strains accumulate glycoproteins with severely truncated N-glycan chains. This hypo-mannosylation did not elicit increased chitin deposition in the cell wall, which in other yeast and fungi is a key compensatory response to cell wall integrity breaches. Thus, C. albicans has evolved an alternate mechanism to adapt to cell wall weakness when N-linked mannan levels are reduced.
Keywords: ANP1; Candida albicans; MNN10; MNN11; MNN9; N-linked glycosylation; OCH1; VAN1; Golgi; cell wall; mannosyltransferase.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


Similar articles
-
Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins.J Bacteriol. 1999 Dec;181(24):7439-48. doi: 10.1128/JB.181.24.7439-7448.1999. J Bacteriol. 1999. PMID: 10601199 Free PMC article.
-
The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans.PLoS Pathog. 2013;9(4):e1003276. doi: 10.1371/journal.ppat.1003276. Epub 2013 Apr 25. PLoS Pathog. 2013. PMID: 23633946 Free PMC article.
-
Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans.J Biol Chem. 2006 Jan 6;281(1):90-8. doi: 10.1074/jbc.M510360200. Epub 2005 Nov 1. J Biol Chem. 2006. PMID: 16263704
-
Fungal Mannosyltransferases as Fitness Attributes and their Contribution to Virulence.Curr Protein Pept Sci. 2017;18(11):1065-1073. doi: 10.2174/1389203717666160813164253. Curr Protein Pept Sci. 2017. PMID: 27526929 Review.
-
N-glycosylation of yeast, with emphasis on Candida albicans.Med Mycol. 2001;39 Suppl 1:75-86. Med Mycol. 2001. PMID: 11800271 Review.
Cited by
-
Histone deacetylase Sir2 promotes the systemic Candida albicans infection by facilitating its immune escape via remodeling the cell wall and maintaining the metabolic activity.mBio. 2024 Jun 12;15(6):e0044524. doi: 10.1128/mbio.00445-24. Epub 2024 Apr 29. mBio. 2024. PMID: 38682948 Free PMC article.
-
Discovery of α-(1→6)-linked mannan structures resembling yeast N-glycan outer chains in Aspergillus fumigatus mycelium.mSphere. 2024 May 29;9(5):e0010024. doi: 10.1128/msphere.00100-24. Epub 2024 Apr 23. mSphere. 2024. PMID: 38651868 Free PMC article.
-
The role of manganese in morphogenesis and pathogenesis of the opportunistic fungal pathogen Candida albicans.PLoS Pathog. 2023 Jun 26;19(6):e1011478. doi: 10.1371/journal.ppat.1011478. eCollection 2023 Jun. PLoS Pathog. 2023. PMID: 37363924 Free PMC article.
-
Mannan is a context-dependent shield that modifies virulence in Nakaseomyces glabratus.Virulence. 2025 Dec;16(1):2491650. doi: 10.1080/21505594.2025.2491650. Epub 2025 Apr 15. Virulence. 2025. PMID: 40233931 Free PMC article.
-
Rapid evolution of an adaptive multicellular morphology of Candida auris during systemic infection.Nat Commun. 2024 Mar 16;15(1):2381. doi: 10.1038/s41467-024-46786-8. Nat Commun. 2024. PMID: 38493178 Free PMC article.
References
-
- Ballou CE. Isolation, characterization and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 1990;185:440–470. - PubMed
-
- Bates S, Hughes HB, Munro CA, Thomas WPH, MacCallum DM, Bertram G, Atrih A, Ferguson MAJ, Brown AJP, Odds FC, et al.Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J Biol Chem. 2006;281(1):90–98. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

