PLK4 is upregulated in prostate cancer and its inhibition reduces centrosome amplification and causes senescence
- PMID: 35333404
- PMCID: PMC9090996
- DOI: 10.1002/pros.24342
PLK4 is upregulated in prostate cancer and its inhibition reduces centrosome amplification and causes senescence
Abstract
Background: Identification of novel molecular target(s) is important for designing newer mechanistically driven approaches for the treatment of prostate cancer (PCa), which is one of the main causes of morbidity and mortality in men. In this study, we determined the role of polo-like kinase 4 (PLK4), which regulates centriole duplication and centrosome amplification (CA), in PCa.
Materials and methods: Employing human PCa tissue microarrays, we assessed the prevalence of CA, correlated with Gleason score, and estimated major causes of CA in PCa (cell doubling vs. centriole overduplication) by staining for mother/mature centrioles. We also assessed PLK4 expression and correlated it with CA in human PCa tissues and cell lines. Further, we determined the effects of PLK4 inhibition in human PCa cells.
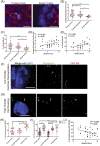
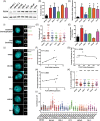
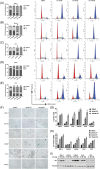
Results: Compared to benign prostate, human PCa demonstrated significantly higher CA, which was also positively correlated with the Gleason score. Further, most cases of CA were found to arise by centriole overduplication rather than cell doubling events (e.g., cytokinesis failure) in PCa. In addition, PLK4 was overexpressed in human PCa cell lines and tumors. Moreover, PLK4 inhibitors CFI-400945 and centrinone-B inhibited cell growth, viability, and colony formation of both androgen-responsive and androgen-independent PCa cell lines. PLK4 inhibition also induced cell cycle arrest and senescence in human PCa cells.
Conclusions: CA is prevalent in PCa and arises predominantly by centriole overduplication as opposed to cell doubling events. Loss of centrioles is cellular stress that can promote senescence and suggests that PLK4 inhibition may be a viable therapeutic strategy in PCa.
Keywords: CFI-400945; PLK4; centriole overduplication; centrosome; prostate cancer.
© 2022 The Authors. The Prostate published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Centriole Overduplication is the Predominant Mechanism Leading to Centrosome Amplification in Melanoma.Mol Cancer Res. 2018 Mar;16(3):517-527. doi: 10.1158/1541-7786.MCR-17-0197. Epub 2018 Jan 12. Mol Cancer Res. 2018. PMID: 29330283 Free PMC article.
-
Cep78 is a new centriolar protein involved in Plk4-induced centriole overduplication.J Cell Sci. 2016 Jul 15;129(14):2713-8. doi: 10.1242/jcs.184093. Epub 2016 May 31. J Cell Sci. 2016. PMID: 27246242
-
Autophosphorylation of polo-like kinase 4 and its role in centriole duplication.Mol Biol Cell. 2010 Feb 15;21(4):547-61. doi: 10.1091/mbc.e09-06-0505. Epub 2009 Dec 23. Mol Biol Cell. 2010. PMID: 20032307 Free PMC article.
-
Role of Polo-like Kinases Plk1 and Plk4 in the Initiation of Centriole Duplication-Impact on Cancer.Cells. 2022 Feb 24;11(5):786. doi: 10.3390/cells11050786. Cells. 2022. PMID: 35269408 Free PMC article. Review.
-
The PLK4-STIL-SAS-6 module at the core of centriole duplication.Biochem Soc Trans. 2016 Oct 15;44(5):1253-1263. doi: 10.1042/BST20160116. Biochem Soc Trans. 2016. PMID: 27911707 Free PMC article. Review.
Cited by
-
Polo-like kinase 4 (Plk4) potentiates anoikis-resistance of p53KO mammary epithelial cells by inducing a hybrid EMT phenotype.Cell Death Dis. 2023 Feb 16;14(2):133. doi: 10.1038/s41419-023-05618-1. Cell Death Dis. 2023. PMID: 36797240 Free PMC article.
-
The yin and yang of chromosomal instability in prostate cancer.Nat Rev Urol. 2024 Jun;21(6):357-372. doi: 10.1038/s41585-023-00845-9. Epub 2024 Feb 2. Nat Rev Urol. 2024. PMID: 38307951 Free PMC article. Review.
-
Aberrant expression of polo-like kinase 4 in renal cell carcinoma: Association with clinicopathological characteristics and long-term survival.Oncol Lett. 2022 Oct 14;24(6):427. doi: 10.3892/ol.2022.13547. eCollection 2022 Dec. Oncol Lett. 2022. PMID: 36311683 Free PMC article.
-
PLK4 is a potential therapeutic target in nonmelanoma skin cancers: Evidence from molecular and in vivo studies.Photochem Photobiol. 2025 Jun 16:10.1111/php.70006. doi: 10.1111/php.70006. Online ahead of print. Photochem Photobiol. 2025. PMID: 40524317
-
Role of PLK4 inhibition in cancer therapy.Cancer Metastasis Rev. 2025 Jun 13;44(2):55. doi: 10.1007/s10555-025-10271-5. Cancer Metastasis Rev. 2025. PMID: 40512236 Review.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7‐33. - PubMed
-
- Pihan GA, Purohit A, Wallace J, et al. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58(17):3974‐3985. - PubMed
-
- Pihan GA, Purohit A, Wallace J, Malhotra R, Liotta L, Doxsey SJ. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 2001;61(5):2212‐2219. - PubMed
-
- Toma MI, Friedrich K, Meyer W, et al. Correlation of centrosomal aberrations with cell differentiation and DNA ploidy in prostate cancer. Anal Quant Cytol Histol. 2010;32(1):1‐10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

