Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis
- PMID: 35334234
- PMCID: PMC9513754
- DOI: 10.1016/S1474-4422(21)00414-2
Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis
Abstract
Amyotrophic lateral sclerosis is a fatal neurodegenerative disease. The discovery of genes associated with amyotrophic lateral sclerosis, commencing with SOD1 in 1993, started fairly gradually. Recent advances in genetic technology have led to the rapid identification of multiple new genes associated with the disease, and to a new understanding of oligogenic and polygenic disease risk. The overlap of genes associated with amyotrophic lateral sclerosis with those of other neurodegenerative diseases is shedding light on the phenotypic spectrum of neurodegeneration, leading to a better understanding of genotype-phenotype correlations. A deepening knowledge of the genetic architecture is allowing the characterisation of the molecular steps caused by various mutations that converge on recurrent dysregulated pathways. Of crucial relevance, mutations associated with amyotrophic lateral sclerosis are amenable to novel gene-based therapeutic options, an approach in use for other neurological illnesses. Lastly, the exposome-the summation of lifetime environmental exposures-has emerged as an influential component for amyotrophic lateral sclerosis through the gene-time-environment hypothesis. Our improved understanding of all these aspects will lead to long-awaited therapies and the identification of modifiable risks factors.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests SAG declares consulting fees from Biogen and ITF Pharma, a patent “Methods for treating amyotrophic lateral sclerosis”, and participation on a Data Safety Monitoring Board for Watermark. OH declares consulting fees from Novartis, Cytokinetics, Denali Pharma, Stitching Foundation, and La Caixa; payment or honoraria from Biogen; participation on a Data Safety Monitoring Board for Accelsiors and steering committee for Cytokinetics; and is Editor-in-Chief for the journal Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. AA-C declares consulting fees from Mitsubishi Tanabe Pharma, Biogen Idec, Cytokinetics, Wave Pharmaceuticals, Apellis, Amylyx, Novartis, and Eli Lilly. AC declares grants from Biogen to his institution, payments or honoraria from Biogen and Amylyx, and participation on a Data Safety Monitoring Board for Ely Lilly and ABScience and advisory board for Mitsubishi Tanabe, Roche, Denali Pharma, Cytokinetics, Biogen, and Amylyx. MCK has an honorary role as President of the Brain Foundation and as Editor-in-Chief of the Journal of Neurology, Neurosurgery and Psychiatry. ELF declares a patent “Methods for treating amyotrophic lateral sclerosis”. MGS declares no competing interests.
Figures

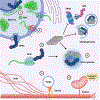
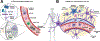
Comment in
-
Amyotrophic lateral sclerosis: new era, new challenges.Lancet Neurol. 2022 May;21(5):400-401. doi: 10.1016/S1474-4422(22)00084-9. Epub 2022 Mar 22. Lancet Neurol. 2022. PMID: 35334235 No abstract available.
-
Amyotrophic lateral sclerosis from genotoxins alone?Lancet Neurol. 2022 Sep;21(9):771-772. doi: 10.1016/S1474-4422(22)00305-2. Lancet Neurol. 2022. PMID: 35963253 No abstract available.
-
Amyotrophic lateral sclerosis from genotoxins alone? - Authors' reply.Lancet Neurol. 2022 Sep;21(9):772. doi: 10.1016/S1474-4422(22)00301-5. Lancet Neurol. 2022. PMID: 35963254 No abstract available.
References
-
- Goutman SA. Diagnosis and Clinical Management of Amyotrophic Lateral Sclerosis and Other Motor Neuron Disorders. Continuum (Minneap Minn). 2017;23(5, Peripheral Nerve and Motor Neuron Disorders):1332–59. - PubMed
Publication types
MeSH terms
Grants and funding
- ALCHALABI-DOBSON/APR14/829-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- R01 TS000327/TS/ATSDR CDC HHS/United States
- AL-CHALABI/APR15/844-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- K23 ES027221/ES/NIEHS NIH HHS/United States
- MR/L501529/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

