Genetic Modification of T Cells for the Immunotherapy of Cancer
- PMID: 35335089
- PMCID: PMC8949949
- DOI: 10.3390/vaccines10030457
Genetic Modification of T Cells for the Immunotherapy of Cancer
Abstract
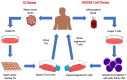
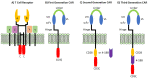
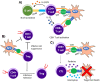
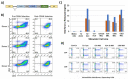
Immunotherapy is a beneficial treatment approach for multiple cancers, however, current therapies are effective only in a small subset of patients. Adoptive cell transfer (ACT) is a facet of immunotherapy where T cells targeting the tumor cells are transferred to the patient with several primary forms, utilizing unmodified or modified T cells: tumor-infiltrating lymphocytes (TIL), genetically modified T cell receptor transduced T cells, and chimeric antigen receptor (CAR) transduced T cells. Many clinical trials are underway investigating the efficacy and safety of these different subsets of ACT, as well as trials that combine one of these subsets with another type of immunotherapy. The main challenges existing with ACT are improving clinical responses and decreasing adverse events. Current research focuses on identifying novel tumor targeting T cell receptors, improving safety and efficacy, and investigating ACT in combination with other immunotherapies.
Keywords: adoptive cell transfer; cancer immunotherapy; chimeric antigen receptors; gene-modified TCR transduced T cells; tumor-infiltrating lymphocytes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Design and implementation of adoptive therapy with chimeric antigen receptor-modified T cells.Immunol Rev. 2014 Jan;257(1):127-44. doi: 10.1111/imr.12139. Immunol Rev. 2014. PMID: 24329794 Free PMC article. Review.
-
Cancer immunotherapy utilizing gene-modified T cells: From the bench to the clinic.Mol Immunol. 2015 Oct;67(2 Pt A):46-57. doi: 10.1016/j.molimm.2014.12.009. Epub 2015 Jan 13. Mol Immunol. 2015. PMID: 25595028 Review.
-
The Role of Immunological Synapse in Predicting the Efficacy of Chimeric Antigen Receptor (CAR) Immunotherapy.Cell Commun Signal. 2020 Aug 25;18(1):134. doi: 10.1186/s12964-020-00617-7. Cell Commun Signal. 2020. PMID: 32843053 Free PMC article. Review.
-
Human effector T cells derived from central memory cells rather than CD8(+)T cells modified by tumor-specific TCR gene transfer possess superior traits for adoptive immunotherapy.Cancer Lett. 2013 Oct 10;339(2):195-207. doi: 10.1016/j.canlet.2013.06.009. Epub 2013 Jun 18. Cancer Lett. 2013. PMID: 23791878
-
Cellular immunotherapy for malignant gliomas.Expert Opin Biol Ther. 2016 Oct;16(10):1265-75. doi: 10.1080/14712598.2016.1214266. Epub 2016 Jul 29. Expert Opin Biol Ther. 2016. PMID: 27434205 Free PMC article. Review.
Cited by
-
Effect of acupuncture and moxibustion on the immune function of patients with malignant tumors: a systematic review and meta-analysis.Front Immunol. 2025 Jul 25;16:1583522. doi: 10.3389/fimmu.2025.1583522. eCollection 2025. Front Immunol. 2025. PMID: 40787472 Free PMC article.
-
Cancer Resistance to Immunotherapy: Comprehensive Insights with Future Perspectives.Pharmaceutics. 2023 Apr 4;15(4):1143. doi: 10.3390/pharmaceutics15041143. Pharmaceutics. 2023. PMID: 37111629 Free PMC article. Review.
-
Present and future of cancer nano-immunotherapy: opportunities, obstacles and challenges.Mol Cancer. 2025 Jan 18;24(1):26. doi: 10.1186/s12943-024-02214-5. Mol Cancer. 2025. PMID: 39827147 Free PMC article. Review.
-
Insights into the Tumor Microenvironment-Components, Functions and Therapeutics.Int J Mol Sci. 2023 Dec 15;24(24):17536. doi: 10.3390/ijms242417536. Int J Mol Sci. 2023. PMID: 38139365 Free PMC article. Review.
-
Progress in immune microenvironment, immunotherapy and prognostic biomarkers in pediatric osteosarcoma.Front Immunol. 2025 Jan 22;16:1548527. doi: 10.3389/fimmu.2025.1548527. eCollection 2025. Front Immunol. 2025. PMID: 39911380 Free PMC article. Review.
References
-
- Allison J.P., McIntyre B.W., Bloch D. Tumor-specific antigen of murine T-lymphoma defined with monoclonal antibody. J. Immunol. 1982;129:2293–2300. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

