Amyloid Cross-Seeding: Mechanism, Implication, and Inhibition
- PMID: 35335141
- PMCID: PMC8955620
- DOI: 10.3390/molecules27061776
Amyloid Cross-Seeding: Mechanism, Implication, and Inhibition
Abstract
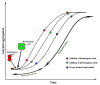
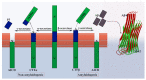
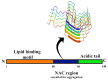
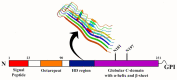
Most neurodegenerative diseases such as Alzheimer's disease, type 2 diabetes, Parkinson's disease, etc. are caused by inclusions and plaques containing misfolded protein aggregates. These protein aggregates are essentially formed by the interactions of either the same (homologous) or different (heterologous) sequences. Several experimental pieces of evidence have revealed the presence of cross-seeding in amyloid proteins, which results in a multicomponent assembly; however, the molecular and structural details remain less explored. Here, we discuss the amyloid proteins and the cross-seeding phenomena in detail. Data suggest that targeting the common epitope of the interacting amyloid proteins may be a better therapeutic option than targeting only one species. We also examine the dual inhibitors that target the amyloid proteins participating in the cross-seeding events. The future scopes and major challenges in understanding the mechanism and developing therapeutics are also considered. Detailed knowledge of the amyloid cross-seeding will stimulate further research in the practical aspects and better designing anti-amyloid therapeutics.
Keywords: aggregation; amyloid proteins; cross-seeding; dual inhibition; fibrillation; protein misfolding diseases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Maji S.K., Perrin M.H., Sawaya M.R., Jessberger S., Vadodaria K., Rissman R.A., Singru P.S., Nilsson K.P., Simon R., Schubert D., et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. - DOI - PMC - PubMed
-
- Fabrizio C., Christopher M.D. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017;86:27–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

