The Use of Long-Read Sequencing Technologies in Infection Control: Horizontal Transfer of a blaCTX-M-27 Containing lncFII Plasmid in a Patient Screening Sample
- PMID: 35336067
- PMCID: PMC8949098
- DOI: 10.3390/microorganisms10030491
The Use of Long-Read Sequencing Technologies in Infection Control: Horizontal Transfer of a blaCTX-M-27 Containing lncFII Plasmid in a Patient Screening Sample
Abstract
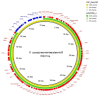
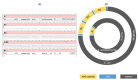
Plasmid transfer is one important mechanism how antimicrobial resistance can spread between different species, contributing to the rise of multidrug resistant bacteria (MDRB) worldwide. Here were present whole genome sequencing (WGS) data of two MDRB isolates, an Escherichia coli and a Klebsiella quasipneumoniae, which were isolated from a single patient. Detailed analysis of long-read sequencing data identified an identical F2:A-:B- lncFII plasmid containing blaCTX-M-27 in both isolates, suggesting horizontal plasmid exchange between the two species. As the plasmid of the E. coli strain carried multiple copies of the resistance cassette, the genomic data correlated with the increased antimicrobial resistance (AMR) detected for this isolate. Our case report demonstrates how long-read sequencing data of MDRB can be used to investigate the role of plasmid mediate resistance in the healthcare setting and explain resistance phenotypes.
Keywords: blaCTX-M-27; long-read sequencing; plasmid transfer.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Novais A., Viana D., Baquero F., Martinez-Botas J., Canton R., Coque T.M. Contribution of IncFII and broad-host IncA/C and IncN plasmids to the local expansion and diversification of phylogroup B2 Escherichia coli ST131 clones carrying blaCTX-M-15 and qnrS1 genes. Antimicrob. Agents Chemother. 2012;56:2763–2766. doi: 10.1128/AAC.06001-11. - DOI - PMC - PubMed
-
- Patino-Navarrete R., Rosinski-Chupin I., Cabanel N., Zongo P.D., Hery M., Oueslati S., Girlich D., Dortet L., Bonnin R.A., Naas T., et al. Specificities and Commonalities of Carbapenemase-Producing Escherichia coli Isolated in France from 2012 to 2015. mSystems. 2022;7:e01169-21. doi: 10.1128/msystems.01169-21. - DOI - PMC - PubMed
-
- Zautner A.E., Bunk B., Pfeifer Y., Sproer C., Reichard U., Eiffert H., Scheithauer S., Gross U., Overmann J., Bohne W. Monitoring microevolution of OXA-48-producing Klebsiella pneumoniae ST147 in a hospital setting by SMRT sequencing. J. Antimicrob. Chemother. 2017;72:2737–2744. doi: 10.1093/jac/dkx216. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

