Genetic Diversity and Relationships of Listeria monocytogenes Serogroup IIa Isolated in Poland
- PMID: 35336111
- PMCID: PMC8951407
- DOI: 10.3390/microorganisms10030532
Genetic Diversity and Relationships of Listeria monocytogenes Serogroup IIa Isolated in Poland
Abstract
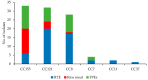
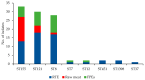
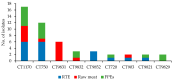
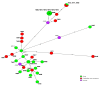
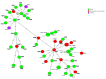
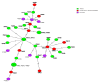
In the present study, 100 L. monocytogenes isolates of serogroup IIa from food and food production environments in Poland were characterized towards the presence of virulence, resistance, and stress response genes using whole-genome sequencing (WGS). The strains were also molecularly typed and compared with multi-locus sequence typing (MLST) and core genome MLST analyses. The present isolates were grouped into 6 sublineages (SLs), with the most prevalent SL155 (33 isolates), SL121 (32 isolates), and SL8 (28 isolates) and classified into six clonal complexes, with the most prevalent CC155 (33 strains), CC121 (32 isolates), and CC8 (28 strains). Furthermore, the strains were grouped to eight sequence types, with the most prevalent ST155 (33 strains), ST121 (30 isolates), and ST8 (28; strains) followed by 60 cgMLST types (CTs). WGS data showed the presence of several virulence genes or putative molecular markers playing a role in pathogenesis of listeriosis and involved in survival of L. monocytogenes in adverse environmental conditions. Some of the present strains were molecularly closely related to L. monocytogenes previously isolated in Poland. The results of the study showed that food and food production environments may be a source of L. monocytogenes of serogroup IIa with pathogenic potential.
Keywords: Listeria monocytogenes; WGS; food; molecular typing; serogroup IIa.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Burall L.S., Grim C.J., Mammel M.K., Datta A.R. Whole genome sequence analysis using JSpecies tool establishes clonal relationships between Listeria monocytogenes strains from epidemiologically unrelated listeriosis outbreaks. PLoS ONE. 2016;11:e0150797. doi: 10.1371/journal.pone.0150797. - DOI - PMC - PubMed
-
- Henri C., Leekitcharoenphon P., Carleton H.A., Radomski N., Kaas R.S., Mariet J.F., Felten A., Aarestrup F.M., Gerner Smidt P., Roussel S., et al. An assessment of different genomic approaches for inferring phylogeny of Listeria monocytogenes. Front. Microbiol. 2017;8:2351. doi: 10.3389/fmicb.2017.02351. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

