The Use and Limitations of the 16S rRNA Sequence for Species Classification of Anaplasma Samples
- PMID: 35336180
- PMCID: PMC8949108
- DOI: 10.3390/microorganisms10030605
The Use and Limitations of the 16S rRNA Sequence for Species Classification of Anaplasma Samples
Abstract
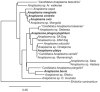
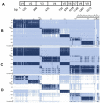
With the advent of cheaper, high-throughput sequencing technologies, the ability to survey biodiversity in previously unexplored niches and geographies has expanded massively. Within Anaplasma, a genus containing several intra-hematopoietic pathogens of medical and economic importance, at least 25 new species have been proposed since the last formal taxonomic organization. Given the obligate intracellular nature of these bacteria, none of these proposed species have been able to attain formal standing in the nomenclature per the International Code of Nomenclature of Prokaryotes rules. Many novel species' proposals use sequence data obtained from targeted or metagenomic PCR studies of only a few genes, most commonly the 16S rRNA gene. We examined the utility of the 16S rRNA gene sequence for discriminating Anaplasma samples to the species level. We find that while the genetic diversity of the genus Anaplasma appears greater than appreciated in the last organization of the genus, caution must be used when attempting to resolve to a species descriptor from the 16S rRNA gene alone. Specifically, genomically distinct species have similar 16S rRNA gene sequences, especially when only partial amplicons of the 16S rRNA are used. Furthermore, we provide key bases that allow classification of the formally named species of Anaplasma.
Keywords: 16S rRNA; Anaplasma; microbiome; species definition; taxonomy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
Cited by
-
Molecular Characterization of Anaplasma spp. in Cattle from Kazakhstan.Pathogens. 2024 Oct 12;13(10):894. doi: 10.3390/pathogens13100894. Pathogens. 2024. PMID: 39452765 Free PMC article.
-
Characteristic gene expression profile of intestinal mucosa early in life promotes bacterial colonization leading to healthy development of the intestinal environment.Sci Rep. 2025 May 26;15(1):18437. doi: 10.1038/s41598-025-03661-w. Sci Rep. 2025. PMID: 40419682 Free PMC article.
-
Detecting genetic gain and loss events in terms of protein domain: Method and implementation.Heliyon. 2024 May 31;10(11):e32103. doi: 10.1016/j.heliyon.2024.e32103. eCollection 2024 Jun 15. Heliyon. 2024. PMID: 38867972 Free PMC article.
-
Anaplasma Species in Africa-A Century of Discovery: A Review on Molecular Epidemiology, Genetic Diversity, and Control.Pathogens. 2023 May 12;12(5):702. doi: 10.3390/pathogens12050702. Pathogens. 2023. PMID: 37242372 Free PMC article. Review.
-
Unraveling the genomic diversity of the Pseudomonas putida group: exploring taxonomy, core pangenome, and antibiotic resistance mechanisms.FEMS Microbiol Rev. 2024 Nov 23;48(6):fuae025. doi: 10.1093/femsre/fuae025. FEMS Microbiol Rev. 2024. PMID: 39390673 Free PMC article. Review.
References
-
- Dumler J.S., Barbet A.F., Bekker C.P.J., Dasch G.A., Palmer G.H., Ray S., Rikihisa Y., Rurangirwa F.R. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: Unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

