Analyses of Long-Term Epidemic Trends and Evolution Characteristics of Haplotype Subtypes Reveal the Dynamic Selection on SARS-CoV-2
- PMID: 35336862
- PMCID: PMC8954678
- DOI: 10.3390/v14030454
Analyses of Long-Term Epidemic Trends and Evolution Characteristics of Haplotype Subtypes Reveal the Dynamic Selection on SARS-CoV-2
Abstract
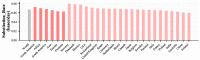
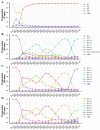
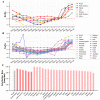
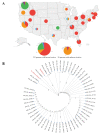
The scale of SARS-CoV-2 infection and death is so enormous that further study of the molecular and evolutionary characteristics of SARS-CoV-2 will help us better understand and respond to SARS-CoV-2 outbreaks. The present study analyzed the epidemic and evolutionary characteristics of haplotype subtypes or regions based on 1.8 million high-quality SARS-CoV-2 genomic data. The estimated ratio of the rates of non-synonymous to synonymous changes (Ka/Ks) in North America and the United States were always more than 1.0, while the Ka/Ks in other continents and countries showed a sharp decline, then a slow increase to 1.0, and a dramatic increase over time. H1 (B.1) with the highest substitution rate has become the most dominant haplotype subtype since March 2020 and has evolved into multiple haplotype subtypes with smaller substitution rates. Many evolutionary characteristics of early SARS-CoV-2, such as H3 being the only early haplotype subtype that existed for the shortest time, the global prevalence of H1 and H1-5 (B.1.1) within a month after being detected, and many high divergent genome sequences early in February 2020, indicate the missing of early SARS-CoV-2 genomic data. SARS-CoV-2 experienced dynamic selection from December 2019 to August 2021 and has been under strong positive selection since May 2021. Its transmissibility and the ability of immune escape may be greatly enhanced over time. This will bring greater challenges to the control of the pandemic.
Keywords: Ka/Ks; SARS-CoV-2; epidemic trends; evolution; haplotype subtypes; substitution rate.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Xi B., Jiang D., Li S., Lon J.R., Bai Y., Lin S., Hu M., Meng Y., Qu Y., Huang Y., et al. AutoVEM: An automated tool to real-time monitor epidemic trends and key mutations in SARS-CoV-2 evolution. Comput. Struct. Biotechnol. J. 2021;19:1976–1985. doi: 10.1016/j.csbj.2021.04.002. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

