Sequence Matching between Hemagglutinin and Neuraminidase through Sequence Analysis Using Machine Learning
- PMID: 35336876
- PMCID: PMC8950662
- DOI: 10.3390/v14030469
Sequence Matching between Hemagglutinin and Neuraminidase through Sequence Analysis Using Machine Learning
Abstract
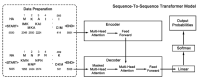
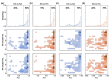
To date, many experiments have revealed that the functional balance between hemagglutinin (HA) and neuraminidase (NA) plays a crucial role in viral mobility, production, and transmission. However, whether and how HA and NA maintain balance at the sequence level needs further investigation. Here, we applied principal component analysis and hierarchical clustering analysis on thousands of HA and NA sequences of A/H1N1 and A/H3N2. We discovered significant coevolution between HA and NA at the sequence level, which is closely related to the type of host species and virus epidemic years. Furthermore, we propose a sequence-to-sequence transformer model (S2STM), which mainly consists of an encoder and a decoder that adopts a multi-head attention mechanism for establishing the mapping relationship between HA and NA sequences. The training results reveal that the S2STM can effectively realize the "translation" from HA to NA or vice versa, thereby building a relationship network between them. Our work combines unsupervised and supervised machine learning methods to identify the sequence matching between HA and NA, which will advance our understanding of IAVs' evolution and also provide a novel idea for sequence analysis methods.
Keywords: hemagglutinin; influenza A viruses; machine learning; neuraminidase; sequence analysis; viral evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures