Advances in HIV-1 Assembly
- PMID: 35336885
- PMCID: PMC8952333
- DOI: 10.3390/v14030478
Advances in HIV-1 Assembly
Abstract
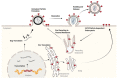
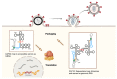
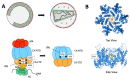
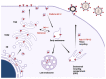
The assembly of HIV-1 particles is a concerted and dynamic process that takes place on the plasma membrane of infected cells. An abundance of recent discoveries has advanced our understanding of the complex sequence of events leading to HIV-1 particle assembly, budding, and release. Structural studies have illuminated key features of assembly and maturation, including the dramatic structural transition that occurs between the immature Gag lattice and the formation of the mature viral capsid core. The critical role of inositol hexakisphosphate (IP6) in the assembly of both the immature and mature Gag lattice has been elucidated. The structural basis for selective packaging of genomic RNA into virions has been revealed. This review will provide an overview of the HIV-1 assembly process, with a focus on recent advances in the field, and will point out areas where questions remain that can benefit from future investigation.
Keywords: Env trafficking; Gag protein; RNA packaging; capsid protein; envelope glycoprotein; inositol hexakisphosphate; matrix protein; maturation; nucleocapsid protein.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Pasquinelli A.E., Ernst R.K., Lund E., Grimm C., Zapp M.L., Rekosh D., Hammarskjold M.L., Dahlberg J.E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

