Prevalence, Diversity and UV-Light Inducibility Potential of Prophages in Bacillus subtilis and Their Possible Roles in Host Properties
- PMID: 35336890
- PMCID: PMC8951512
- DOI: 10.3390/v14030483
Prevalence, Diversity and UV-Light Inducibility Potential of Prophages in Bacillus subtilis and Their Possible Roles in Host Properties
Abstract
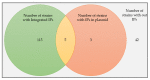
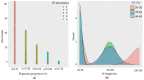
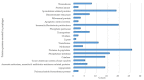
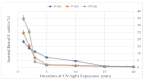
Bacillus subtilis is an important bacterial species due to its various industrial, medicinal, and agricultural applications. Prophages are known to play vital roles in host properties. Nevertheless, studies on the prophages and temperate phages of B. subtilis are relatively limited. In the present study, an in silico analysis was carried out in sequenced B. subtilis strains to investigate their prevalence, diversity, insertion sites, and potential roles. In addition, the potential for UV induction and prevalence was investigated. The in silico prophage analysis of 164 genomes of B. subtilis strains revealed that 75.00% of them contained intact prophages that exist as integrated and/or plasmid forms. Comparative genomics revealed the rich diversity of the prophages distributed in 13 main clusters and four distinct singletons. The analysis of the putative prophage proteins indicated the involvement of prophages in encoding the proteins linked to the immunity, bacteriocin production, sporulation, arsenate, and arsenite resistance of the host, enhancing its adaptability to diverse environments. An induction study in 91 B. subtilis collections demonstrated that UV-light treatment was instrumental in producing infective phages in 18.68% of them, showing a wide range of host specificity. The high prevalence and inducibility potential of the prophages observed in this study implies that prophages may play vital roles in the B. subtilis host.
Keywords: Bacillus subtilis; bacteriophages; diversity; in silico prophage analysis; induction; insertion sites; prevalence; prophages.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Putative plasmid prophages of Bacillus cereus sensu lato may hold the key to undiscovered phage diversity.Sci Rep. 2021 Apr 7;11(1):7611. doi: 10.1038/s41598-021-87111-3. Sci Rep. 2021. PMID: 33828147 Free PMC article.
-
Ecology of prophage-like elements in Bacillus subtilis at global and local geographical scales.Cell Rep. 2025 Jan 28;44(1):115197. doi: 10.1016/j.celrep.2024.115197. Epub 2025 Jan 10. Cell Rep. 2025. PMID: 39798088
-
Prevalence and Diversity Analysis of Candidate Prophages to Provide An Understanding on Their Roles in Bacillus Thuringiensis.Viruses. 2019 Apr 25;11(4):388. doi: 10.3390/v11040388. Viruses. 2019. PMID: 31027262 Free PMC article.
-
Research progress of prophages.Yi Chuan. 2021 Mar 16;43(3):240-248. doi: 10.16288/j.yczz.20-355. Yi Chuan. 2021. PMID: 33724208 Review.
-
Comparative genomics of phages and prophages in lactic acid bacteria.Antonie Van Leeuwenhoek. 2002 Aug;82(1-4):73-91. Antonie Van Leeuwenhoek. 2002. PMID: 12369206 Review.
Cited by
-
Genomic diversity and comprehensive taxonomical classification of 61 Bacillus subtilis group member infecting bacteriophages, and the identification of ortholog taxonomic signature genes.BMC Genomics. 2022 Dec 16;23(1):835. doi: 10.1186/s12864-022-09055-w. BMC Genomics. 2022. PMID: 36526963 Free PMC article.
-
Novel MRSA-targeting phage MetB16: Genomic features, structural insights, and therapeutic applications.Turk J Biol. 2025 Feb 14;49(3):292-308. doi: 10.55730/1300-0152.2746. eCollection 2025. Turk J Biol. 2025. PMID: 40678418 Free PMC article.
References
-
- Colavecchio A., D’Souza Y., Tompkins E., Jeukens J., Freschi L., Emond-Rheault J.-G., Kukavica-Ibrulj I., Boyle B., Bekal S., Tamber S. Prophage integrase typing is a useful indicator of genomic diversity in Salmonella enterica. Front. Microbiol. 2017;8:1283. doi: 10.3389/fmicb.2017.01283. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

