Early Genomic, Epidemiological, and Clinical Description of the SARS-CoV-2 Omicron Variant in Mexico City
- PMID: 35336952
- PMCID: PMC8950183
- DOI: 10.3390/v14030545
Early Genomic, Epidemiological, and Clinical Description of the SARS-CoV-2 Omicron Variant in Mexico City
Abstract
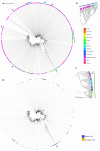
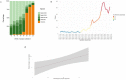
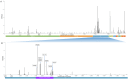
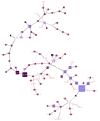
Omicron is the most mutated SARS-CoV-2 variant-a factor that can affect transmissibility, disease severity, and immune evasiveness. Its genomic surveillance is important in cities with millions of inhabitants and an economic center, such as Mexico City. Results. From 16 November to 31 December 2021, we observed an increase of 88% in Omicron prevalence in Mexico City. We explored the R346K substitution, prevalent in 42% of Omicron variants, known to be associated with immune escape by monoclonal antibodies. In a phylogenetic analysis, we found several independent exchanges between Mexico and the world, and there was an event followed by local transmission that gave rise to most of the Omicron diversity in Mexico City. A haplotype analysis revealed that there was no association between haplotype and vaccination status. Among the 66% of patients who have been vaccinated, no reported comorbidities were associated with Omicron; the presence of odynophagia and the absence of dysgeusia were significant predictor symptoms for Omicron, and the RT-qPCR Ct values were lower for Omicron. Conclusions. Genomic surveillance is key to detecting the emergence and spread of SARS-CoV-2 variants in a timely manner, even weeks before the onset of an infection wave, and can inform public health decisions and detect the spread of any mutation that may affect therapeutic efficacy.
Keywords: Omicron variant; R346K; SARS-CoV-2; dysgeusia; haplotype analysis; odynophagia; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

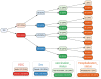
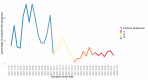
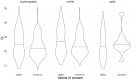
References
-
- Khandia R., Singhal S., Alqahtani T., Kamal M.A., El-Shall N.A., Nainu F., Desingu P.A., Dhama K. Emergence of SARS-CoV-2 Omicron (B.1.1.529) Variant, Salient Features, High Global Health Concerns and Strategies to Counter It amid Ongoing COVID-19 Pandemic. Environ. Res. 2022;209:112816. doi: 10.1016/j.envres.2022.112816. - DOI - PMC - PubMed
-
- Sifuentes-Osornio J., Angulo-Guerrero O., De-Anda-Jáuregui G., Díaz-De-León-Santiago J.L., Hernández-Lemus E., Benítez-Pérez H., Herrera L.A., López-Arellano O., Revuelta-Herrera A., Rosales-Tapia A.R., et al. Probability of Hospitalization and Death among COVID-19 Patients with Comorbidity during Outbreaks Occurring in Mexico City. Medrxiv. 2021 doi: 10.1101/2021.12.07.21267287. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

