RNA Structural Requirements for Nucleocapsid Protein-Mediated Extended Dimer Formation
- PMID: 35337013
- PMCID: PMC8953772
- DOI: 10.3390/v14030606
RNA Structural Requirements for Nucleocapsid Protein-Mediated Extended Dimer Formation
Abstract
Retroviruses package two copies of their genomic RNA (gRNA) as non-covalently linked dimers. Many studies suggest that the retroviral nucleocapsid protein (NC) plays an important role in gRNA dimerization. The upper part of the L3 RNA stem-loop in the 5' leader of the avian leukosis virus (ALV) is converted to the extended dimer by ALV NC. The L3 hairpin contains three stems and two internal loops. To investigate the roles of internal loops and stems in the NC-mediated extended dimer formation, we performed site-directed mutagenesis, gel electrophoresis, and analysis of thermostability of dimeric RNAs. We showed that the internal loops are necessary for efficient extended dimer formation. Destabilization of the lower stem of L3 is necessary for RNA dimerization, although it is not involved in the linkage structure of the extended dimer. We found that NCs from ALV, human immunodeficiency virus type 1 (HIV-1), and Moloney murine leukemia virus (M-MuLV) cannot promote the formation of the extended dimer when the apical stem contains ten consecutive base pairs. Five base pairs correspond to the maximum length for efficient L3 dimerization induced by the three NCs. L3 dimerization was less efficient with M-MuLV NC than with ALV NC and HIV-1 NC.
Keywords: HIV-1; RNA dimerization; RNA secondary structure; Rous sarcoma virus (RSV); nucleocapsid protein; retrovirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
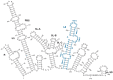

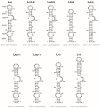

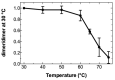



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

