A BioID-Derived Proximity Interactome for SARS-CoV-2 Proteins
- PMID: 35337019
- PMCID: PMC8951556
- DOI: 10.3390/v14030611
A BioID-Derived Proximity Interactome for SARS-CoV-2 Proteins
Abstract
The novel coronavirus SARS-CoV-2 is responsible for the ongoing COVID-19 pandemic and has caused a major health and economic burden worldwide. Understanding how SARS-CoV-2 viral proteins behave in host cells can reveal underlying mechanisms of pathogenesis and assist in development of antiviral therapies. Here, the cellular impact of expressing SARS-CoV-2 viral proteins was studied by global proteomic analysis, and proximity biotinylation (BioID) was used to map the SARS-CoV-2 virus-host interactome in human lung cancer-derived cells. Functional enrichment analyses revealed previously reported and unreported cellular pathways that are associated with SARS-CoV-2 proteins. We have established a website to host the proteomic data to allow for public access and continued analysis of host-viral protein associations and whole-cell proteomes of cells expressing the viral-BioID fusion proteins. Furthermore, we identified 66 high-confidence interactions by comparing this study with previous reports, providing a strong foundation for future follow-up studies. Finally, we cross-referenced candidate interactors with the CLUE drug library to identify potential therapeutics for drug-repurposing efforts. Collectively, these studies provide a valuable resource to uncover novel SARS-CoV-2 biology and inform development of antivirals.
Keywords: BioID; COVID-19; SARS-CoV-2; TurboID; interactome; proximity labeling.
Conflict of interest statement
Sanford Research has licensed BioID reagents to BioFront Technologies. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
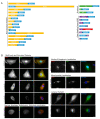


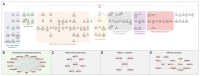

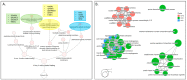
Update of
-
A BioID-derived proximity interactome for SARS-CoV-2 proteins.bioRxiv [Preprint]. 2021 Sep 21:2021.09.17.460814. doi: 10.1101/2021.09.17.460814. bioRxiv. 2021. Update in: Viruses. 2022 Mar 15;14(3):611. doi: 10.3390/v14030611. PMID: 34580671 Free PMC article. Updated. Preprint.
References
-
- Orrù G., Bertelloni D., Diolaiuti F., Mucci F., Di Giuseppe M., Biella M., Gemignani A., Ciacchini R., Conversano C. Long-COVID Syndrome? A Study on the Persistence of Neurological, Psychological and Physiological Symptoms. Healthcare. 2021;9:575. doi: 10.3390/healthcare9050575. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- R35GM126949/NH/NIH HHS/United States
- P30 CA030199/CA/NCI NIH HHS/United States
- P20 GM103620/GM/NIGMS NIH HHS/United States
- U19 AI135990/AI/NIAID NIH HHS/United States
- U24CA184427/GM/NIGMS NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- R01 HG009979/HG/NHGRI NIH HHS/United States
- U24CA184427/CA/NCI NIH HHS/United States
- P41-GM103504/GM/NIGMS NIH HHS/United States
- R01HG009979/HG/NHGRI NIH HHS/United States
- P20GM103620/NH/NIH HHS/United States
- P41 GM103504/GM/NIGMS NIH HHS/United States
- R35 GM126949/GM/NIGMS NIH HHS/United States
- U19 AI135972/AI/NIAID NIH HHS/United States
- U24 CA184427/CA/NCI NIH HHS/United States
- P20 GM103548/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

