Investigation of the Low-Populated Excited States of the HIV-1 Nucleocapsid Domain
- PMID: 35337039
- PMCID: PMC8950434
- DOI: 10.3390/v14030632
Investigation of the Low-Populated Excited States of the HIV-1 Nucleocapsid Domain
Abstract
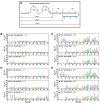
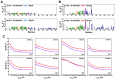
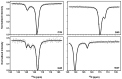
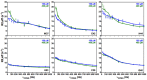
The nucleocapsid domain (NCd), located at the C-terminus of the HIV-1 Gag protein, is involved in numerous stages of the replication cycle, such as the packaging of the viral genome and reverse transcription. It exists under different forms through the viral life cycle, depending on the processing of Gag by the HIV-1 protease. NCd is constituted of two adjacent zinc knuckles (ZK1 and ZK2), separated by a flexible linker and flanked by disordered regions. Here, conformational equilibria between a major and two minor states were highlighted exclusively in ZK2, by using CPMG and CEST NMR experiments. These minor states appear to be temperature dependent, and their populations are highest at physiological temperature. These minor states are present both in NCp7, the mature form of NCd, and in NCp9 and NCp15, the precursor forms of NCd, with increased populations. The role of these minor states in the targeting of NCd by drugs and its binding properties is discussed.
Keywords: CEST; CPMG; HIV-1; NCp15; NCp7; NCp9; NMR; dark-state; dynamic; low-populated state; nucleocapsid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

