A synthetic lipopeptide targeting top-priority multidrug-resistant Gram-negative pathogens
- PMID: 35338128
- PMCID: PMC8956739
- DOI: 10.1038/s41467-022-29234-3
A synthetic lipopeptide targeting top-priority multidrug-resistant Gram-negative pathogens
Abstract
The emergence of multidrug-resistant (MDR) Gram-negative pathogens is an urgent global medical challenge. The old polymyxin lipopeptide antibiotics (polymyxin B and colistin) are often the only therapeutic option due to resistance to all other classes of antibiotics and the lean antibiotic drug development pipeline. However, polymyxin B and colistin suffer from major issues in safety (dose-limiting nephrotoxicity, acute toxicity), pharmacokinetics (poor exposure in the lungs) and efficacy (negligible activity against pulmonary infections) that have severely limited their clinical utility. Here we employ chemical biology to systematically optimize multiple non-conserved positions in the polymyxin scaffold, and successfully disconnect the therapeutic efficacy from the toxicity to develop a new synthetic lipopeptide, structurally and pharmacologically distinct from polymyxin B and colistin. This resulted in the clinical candidate F365 (QPX9003) with superior safety and efficacy against lung infections caused by top-priority MDR pathogens Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae.
© 2022. The Author(s).
Conflict of interest statement
J.Li., T.V., R.L.N., K.D.R., P.E.T. are listed as inventors on the patent application WO2015149131 ‘Polymyxin Derivatives as Antimicrobial Compounds’ which covers F365 and the other lipopeptides listed in this manuscript and has been licensed to Qpex Biopharma. O.L., S.J.H., D.C.G., and M.N.D are employees of Qpex Biopharma and hold equity in that company. All other authors declare no competing interests.
Figures
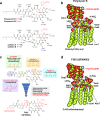

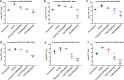
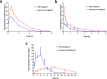
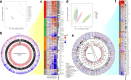
References
-
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. (World Health Organization, 2017).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

