Functional mapping of PHF6 complexes in chromatin remodeling, replication dynamics, and DNA repair
- PMID: 35338774
- PMCID: PMC9185155
- DOI: 10.1182/blood.2021014103
Functional mapping of PHF6 complexes in chromatin remodeling, replication dynamics, and DNA repair
Abstract
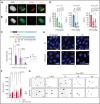
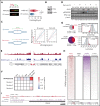
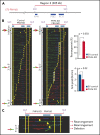
The Plant Homeodomain 6 gene (PHF6) encodes a nucleolar and chromatin-associated leukemia tumor suppressor with proposed roles in transcription regulation. However, specific molecular mechanisms controlled by PHF6 remain rudimentarily understood. Here we show that PHF6 engages multiple nucleosome remodeling protein complexes, including nucleosome remodeling and deacetylase, SWI/SNF and ISWI factors, the replication machinery and DNA repair proteins. Moreover, after DNA damage, PHF6 localizes to sites of DNA injury, and its loss impairs the resolution of DNA breaks, with consequent accumulation of single- and double-strand DNA lesions. Native chromatin immunoprecipitation sequencing analyses show that PHF6 specifically associates with difficult-to-replicate heterochromatin at satellite DNA regions enriched in histone H3 lysine 9 trimethyl marks, and single-molecule locus-specific analyses identify PHF6 as an important regulator of genomic stability at fragile sites. These results extend our understanding of the molecular mechanisms controlling hematopoietic stem cell homeostasis and leukemia transformation by placing PHF6 at the crossroads of chromatin remodeling, replicative fork dynamics, and DNA repair.
© 2022 by The American Society of Hematology.
Figures



Comment in
-
Linking epigenome regulation with DNA repair.Blood. 2022 Jun 9;139(23):3356-3357. doi: 10.1182/blood.2022016176. Blood. 2022. PMID: 35679076 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

