Epstein-Barr Virus in Burkitt Lymphoma in Africa Reveals a Limited Set of Whole Genome and LMP-1 Sequence Patterns: Analysis of Archival Datasets and Field Samples From Uganda, Tanzania, and Kenya
- PMID: 35340265
- PMCID: PMC8948429
- DOI: 10.3389/fonc.2022.812224
Epstein-Barr Virus in Burkitt Lymphoma in Africa Reveals a Limited Set of Whole Genome and LMP-1 Sequence Patterns: Analysis of Archival Datasets and Field Samples From Uganda, Tanzania, and Kenya
Abstract
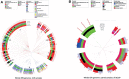
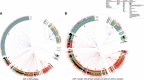
Epstein-Barr virus (EBV) is associated with endemic Burkitt lymphoma (eBL), but the contribution of EBV variants is ill-defined. Studies of EBV whole genome sequences (WGS) have identified phylogroups that appear to be distinct for Asian versus non-Asian EBV, but samples from BL or Africa, where EBV was first discovered, are under-represented. We conducted a phylogenetic analysis of EBV WGS and LMP-1 sequences obtained primarily from BL patients in Africa and representative non-African EBV from other conditions or regions using data from GenBank, Sequence Read Archive, or Genomic Data Commons for the Burkitt Lymphoma Genome Sequencing Project (BLGSP) to generate data to support the use of a simpler biomarker of geographic or phenotypic associations. We also investigated LMP-1 patterns in 414 eBL cases and 414 geographically matched controls in the Epidemiology of Burkitt Lymphoma in East African children and minors (EMBLEM) study using LMP-1 PCR and Sanger sequencing. Phylogenetic analysis revealed distinct genetic patterns of African versus Asian EBV sequences. We identified 281 single nucleotide variations (SNVs) in LMP-1 promoter and coding region, which formed 12 unique patterns (A to L). Nine patterns (A, AB, C, D, F, I, J, K and L) predominated in African EBV, of which four were found in 92% of BL samples (A, AB, D, and H). Predominant patterns were B and G in Asia and H in Europe. EBV positivity in peripheral blood was detected in 95.6% of EMBLEM eBL cases versus 79.2% of the healthy controls (odds ratio [OR] =3.83; 95% confidence interval 2.06-7.14). LMP-1 was successfully sequenced in 66.7% of the EBV DNA positive cases but in 29.6% of the controls (ORs ranging 5-11 for different patterns). Four LMP-1 patterns (A, AB, D, and K) were detected in 63.1% of the cases versus 27.1% controls (ORs ranges: 5.58-11.4). Dual strain EBV infections were identified in WGS and PCR-Sanger data. In conclusion, EBV from Africa is phylogenetically separate from EBV in Asia. Genetic diversity in LMP-1 formed 12 patterns, which showed promising geographic and phenotypic associations. Presence of multiple strain infection should be considered in efforts to refine or improve EBV markers of ancestry or phenotype.
Lay summary: Epstein-Barr virus (EBV) infection, a ubiquitous infection, contributes to the etiology of both Burkitt Lymphoma (BL) and nasopharyngeal carcinoma, yet their global distributions vary geographically with no overlap. Genomic variation in EBV is suspected to play a role in the geographical patterns of these EBV-associated cancers, but relatively few EBV samples from BL have been comprehensively studied. We sought to compare phylogenetic patterns of EBV genomes obtained from BL samples in Africa and from tumor and non-tumor samples from elsewhere. We concluded that EBV obtained from BL in Africa is genetically separate from EBV in Asia. Through comprehensive analysis of nucleotide variations in EBV's LMP-1 gene, we describe 12 LMP-1 patterns, two of which (B and G) were found mostly in Asia. Four LMP-1 patterns (A, AB, D, and F) accounted for 92% of EBVs sequenced from BL in Africa. Our results identified extensive diversity of EBV, but BL in Africa was associated with a limited number of variants identified, which were different from those identified in Asia. Further research is needed to optimize the use of PCR and sequencing to study LMP-1 diversity for classification of EBV variants and for use in epidemiologic studies to characterize geographic and/or phenotypic associations of EBV variants with EBV-associated malignancies, including eBL.
Keywords: Burkitt lymphoma; EBV variants; East Africa; Epstein-Barr virus; LMP-1 patterns; childhood cancer; epidemiology.
Copyright © 2022 Liao, Liu, Chin, Li, Hung, Tsai, Otim, Legason, Ogwang, Reynolds, Kerchan, Tenge, Were, Kuremu, Wekesa, Masalu, Kawira, Ayers, Pfeiffer, Bhatia, Goedert, Lo and Mbulaiteye.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Proceedings of the IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. In: Epstein-Barr Virus and Kaposi's Sarcoma Herpesvirus/Human Herpesvirus 8, vol. 70. Lyon, France: IARC Monogr Eval Carcinog Risks Hum. p. 1–492. 17-24 June 1997. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

