Normalized Ribo-Seq for Quantifying Absolute Global and Specific Changes in Translation
- PMID: 35340296
- PMCID: PMC8899559
- DOI: 10.21769/BioProtoc.4323
Normalized Ribo-Seq for Quantifying Absolute Global and Specific Changes in Translation
Abstract
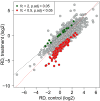
Ribosome profiling (Ribo-Seq) is a highly sensitive method to quantify ribosome occupancies along individual mRNAs on a genome-wide scale. Hereby, ribosome-protected fragments (= footprints) are generated by nuclease digestion, isolated, and sequenced together with the corresponding randomly fragmented input samples, to determine ribosome densities (RD). For library preparation, equal amounts of total RNA are used. Subsequently, all transcript fragments are subjected to linker ligation, cDNA synthesis, and PCR amplification. Importantly, the number of reads obtained for every transcript in input and footprint samples during sequencing depends on sequencing depth and library size, as well as the relative abundance of the transcript in the sample. However, the information pertaining to the absolute amount of input and footprint sequences is lost during sample preparation, hence ruling out any conclusion whether translation is generally suppressed or activated in one condition over the other. Therefore, the RD fold-changes determined for individual genes do not reflect absolute regulation, but have to be interpreted as relative to bulk mRNA translation. Here, we modified the original ribosome profiling protocol that was first established by Ingolia et al. (2009), by adding small amounts of yeast lysate to the mammalian lysates of interest as a spike-in. This allows us to not only detect changes in the RD of specific transcripts relative to each other, but also to simultaneously measure global differences in RD (normalized ribosome density values) between samples. Graphic abstract: Global changes in translation efficiency can be detected with polysome profiling, where the proportion of polysomal ribosomes is interpreted as a proxy for ribosome density (RD) on bulk mRNA. Ribo-Seq measures changes in RD of specific mRNAs relative to bulk mRNA. The addition of a yeast-lysate, as a spike-in for normalization of read counts, allows for an absolute measurement of changes in RD.
Keywords: Normalized ribosome density; Regulation of translation; Ribo-seq; Ribosome profiling; Spike-in.
Copyright © 2022 The Authors; exclusive licensee Bio-protocol LLC.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures



References
-
- Aviner R., Geiger T. and Elroy-Stein O.(2014). Genome-wide identification and quantification of protein synthesis in cultured cells and whole tissues by puromycin-associated nascent chain proteomics(PUNCH-P). Nat Protoc 9(4): 751-760. - PubMed
-
- Cattie D. J., Richardson C. E., Reddy K. C., Ness-Cohn E. M., Droste R., Thompson M. K., Gilbert W. V. and Kim D. H.(2016). Mutations in Nonessential eIF3k and eIF3l Genes Confer Lifespan Extension and Enhanced Resistance to ER Stress in Caenorhabditis elegans . PLoS Genet 12(9): e1006326. - PMC - PubMed
-
- Dieterich D. C., Lee J. J., Link A. J., Graumann J., Tirrell D. A. and Schuman E. M.(2007). Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc 2(3): 532-540. - PubMed
-
- Friedrich U. A., Zedan M., Hessling B., Fenzl K., Gillet L., Barry J., Knop M., Kramer G. and Bukau B.(2021). Nα-terminal acetylation of proteins by NatA and NatB serves distinct physiological roles in Saccharomyces cerevisiae . Cell Rep 34(5): 108711. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

