A bibliometric review of peripartum cardiomyopathy compared to other cardiomyopathies using artificial intelligence and machine learning
- PMID: 35340600
- PMCID: PMC8921361
- DOI: 10.1007/s12551-022-00933-x
A bibliometric review of peripartum cardiomyopathy compared to other cardiomyopathies using artificial intelligence and machine learning
Abstract
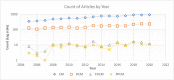
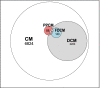
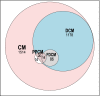
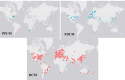
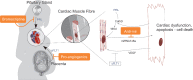
As developments in artificial intelligence and machine learning become more widespread in healthcare, their potential to transform clinical outcomes also increases. Peripartum cardiomyopathy is a rare and poorly-characterised condition that presents as heart failure in the last trimester prior to delivery or within 5-6 months postpartum. The lack of a definitive understanding of the molecular causes and clinical progress of this condition suggests that bibliometrics will be well-suited to creating new insights into this serious clinical problem. We examine similarities and differences between peripartum and its closely related familial dilated cardiomyopathy and idiopathic dilated cardiomyopathy. Using PubMed as the source of bibliometric data, we apply artificial intelligence-supported natural language processing to compare extracted data and genes association with these cardiomyopathies. Gene data were enhanced with additional metadata from third-party datasets and then analysed for their impact and specificity for peripartum cardiomyopathy. Artificial intelligence identified 14 genes that distinguished peripartum from both dilated and familial dilated cardiomyopathy. They are as follows: CTSD, RLN2, MMP23B*, SLC17A5, ST2*, PTHLH, CFH*, CFI, GPT, MR1, Rln1, SRI, STAT5A* and THBD. We then used the Human Protein Atlas website that uses affinity-purified rabbit polyclonal antibodies to identify genes that are expressed at the protein level (bold), or as RNA transcripts (*) in healthy human left ventricles. Additional analysis focussed on the full set of peripartum genes on linkage and specificity to cardiomyopathy yielded a different set of thirteen genes (bold font indicates those expressed in cardiomyocytes: PRL, RLN2, PLN, ST2, CTSD, F2, ACE, STAT3, TTN, SPP1, LGALS3, miR-146a, GNB3, SRI). This type of analysis can highlight new avenues for research, aimed at improving genomics-driven peripartum cardiomyopathy diagnosis as well as potential pathological and clinical sub-classification. We expect that this will allow for future improvements in identification, treatment and management of this condition. The first step in the application of these bibliometric-based artificial intelligence methods is to understand the current knowledge, and it is the aim of this paper to show how this might be achieved.
Keywords: Artificial intelligence; Bibliometrics; Genomics; Machine learning; Peripartum cardiomyopathy.
© International Union for Pure and Applied Biophysics (IUPAB) and Springer-Verlag GmbH Germany, part of Springer Nature 2022.
Conflict of interest statement
Conflict of interestMG, HL, DV, and ST are employees of 23 Strands Pty Ltd (Australia), a privately held company, but their employment does not alter the authors’ adherence to the publication policies.
Figures





References
-
- Adedinsewo DA, Johnson PW, Douglass EJ, Attia IZ, Phillips SD, Goswami RM, Yamani MH, Connolly HM, Rose CH, Sharpe EE, Blauwet L, Lopez-Jimenez F, Friedman PA, Carter RE, Noseworthy PA. Detecting cardiomyopathies in pregnancy and the postpartum period with an electrocardiogram-based deep learning model. Eur Heart J Digit Health. 2021;2:586–596. doi: 10.1093/ehjdh/ztab078. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

